Publications
Preprints
Johansson C,
Johansson M,
Carreras Puigvert J,
Spjuth O,
and Jansson E.
Integrating Cell Painting and Thermal Proteome Profiling for Inference of Targets and Mechanism of Action
bioRxiv. 2025.05.30.657006 (2025). DOI: 10.1101/2025.05.30.657006
Integrating Cell Painting and Thermal Proteome Profiling for Inference of Targets and Mechanism of Action
bioRxiv. 2025.05.30.657006 (2025). DOI: 10.1101/2025.05.30.657006
Seal S,
Dee W,
Shah A,
Zhang A,
Titterton K,
Cabrera AA,
Boiko D,
Beatson A,
Carreras Puigvert J,
Singh S,
Spjuth S,
Bender A,
Carpenter AE.
Small molecule bioactivity benchmarks are often well-predicted by counting cells
bioRxiv. 2025.04.27.650853 (2025). DOI: 10.1101/2025.04.27.650853
Small molecule bioactivity benchmarks are often well-predicted by counting cells
bioRxiv. 2025.04.27.650853 (2025). DOI: 10.1101/2025.04.27.650853
Forsgren E,
Rietdijk J,
Holmberg D,
Johansson M,
Carreras-Puigvert J,
Trygg J,
Lovell G,
Spjuth O,
and Jonsson P.
Label-Free Live-Cell Imaging improves Mode of Action Classification
bioRxiv. 2025.04.22.649936 (2025). DOI: 10.1101/2025.04.22.649936
Label-Free Live-Cell Imaging improves Mode of Action Classification
bioRxiv. 2025.04.22.649936 (2025). DOI: 10.1101/2025.04.22.649936
Ringers C,
Holmberg D,
Flobak Å,
Georgieva P,
Jarvius M,
Johansson M,
Larsson A,
Rosén D,
Seashore–Ludlow B,
Visnes T,
Carreras Puigvert J,
and Spjuth O.
High-content morphological profiling by Cell Painting in 3D spheroids
bioRxiv. 2025.02.05.636642 (2025). DOI: 10.1101/2025.02.05.636642
High-content morphological profiling by Cell Painting in 3D spheroids
bioRxiv. 2025.02.05.636642 (2025). DOI: 10.1101/2025.02.05.636642
Frey B,
Holmberg D,
Bystrom P,
Bergman E,
Georgiev P,
Johansson M,
Hennig P,
Rietdijk J,
Rosen D,
Carreras-Puigvert J,
and Spjuth O.
Single-cell morphological profiling reveals insights into cell death
bioRxiv. 2025.01.15.633042 (2025). DOI: 10.1101/2025.01.15.633042
Single-cell morphological profiling reveals insights into cell death
bioRxiv. 2025.01.15.633042 (2025). DOI: 10.1101/2025.01.15.633042
Söderberg O,
Wenson L,
Heldin J,
Martin M,
Erbilgin Y,
Salman B,
Schaal W,
Sandbaumhüter F,
Jansson E,
Chen X,
Davidsson A,
Stenerlöw B,
Spjuth O.
Precise mapping of single-stranded DNA breaks by using an engineered, error-prone DNA polymerase for sequence-templated erroneous end-labelling
Research Square. (2024). DOI: 10.21203/rs.3.rs-5071189/v1
Precise mapping of single-stranded DNA breaks by using an engineered, error-prone DNA polymerase for sequence-templated erroneous end-labelling
Research Square. (2024). DOI: 10.21203/rs.3.rs-5071189/v1
Fagerholm U,
Hellberg S,
Alvarsson J,
Ekmefjord M and Spjuth O.
Comparing Lipinskis Rule of 5 and Machine Learning Based Prediction of Fraction Absorbed for Assessing Oral Absorption in Humans
bioRxiv. 2024.08.20.608791 (2024). DOI: 10.1101/2024.08.20.608791
Comparing Lipinskis Rule of 5 and Machine Learning Based Prediction of Fraction Absorbed for Assessing Oral Absorption in Humans
bioRxiv. 2024.08.20.608791 (2024). DOI: 10.1101/2024.08.20.608791
Seal S,
Trapotsi MA,
Subramanian V,
Spjuth O,
Greene N,
and Bender A.
PKSmart: An Open-Source Computational Model to Predict in vivo Pharmacokinetics of Small Molecules
bioRxiv. 2024.02.02.578658 (2024). DOI: 10.1101/2024.02.02.578658
PKSmart: An Open-Source Computational Model to Predict in vivo Pharmacokinetics of Small Molecules
bioRxiv. 2024.02.02.578658 (2024). DOI: 10.1101/2024.02.02.578658
Fagerholm U,
Hellberg S,
Alvarsson J,
Spjuth O
Predicting the Influence of Fat Food Intake on the Absorption and Systemic Exposure of Small Drugs using ANDROMEDA by Prosilico Software
bioRxiv. 2022.12.05.519072 (2022). DOI: 10.1101/2022.12.05.519072
Predicting the Influence of Fat Food Intake on the Absorption and Systemic Exposure of Small Drugs using ANDROMEDA by Prosilico Software
bioRxiv. 2022.12.05.519072 (2022). DOI: 10.1101/2022.12.05.519072
Fagerholm U,
Hellberg S,
Alvarsson J,
Spjuth O
Prediction and Classification of the Uptake and Disposition of Antidepressants and New CNS-Active Drugs in the Human Brain using the ANDROMEDA by Prosilico Software and Brainavailability-Matrix
bioRxiv. 2022.09.28.509936 (2022). DOI: 10.1101/2022.09.28.509936
Prediction and Classification of the Uptake and Disposition of Antidepressants and New CNS-Active Drugs in the Human Brain using the ANDROMEDA by Prosilico Software and Brainavailability-Matrix
bioRxiv. 2022.09.28.509936 (2022). DOI: 10.1101/2022.09.28.509936
Fagerholm U,
Hellberg S,
Alvarsson J,
Spjuth O
Prediction of Biopharmaceutical Characteristics of PROTACs using the ANDROMEDA by Prosilico Software
bioRxiv. 2022.09.22.509053 (2022). DOI: 10.1101/2022.09.22.509053
Prediction of Biopharmaceutical Characteristics of PROTACs using the ANDROMEDA by Prosilico Software
bioRxiv. 2022.09.22.509053 (2022). DOI: 10.1101/2022.09.22.509053
Lapins M,
Spjuth O
Evaluation of gene expression and phenotypic profiling data as quantitative descriptors for predicting drug targets and mechanisms of action
bioRxiv. (2019). DOI: 10.1101/580654
Evaluation of gene expression and phenotypic profiling data as quantitative descriptors for predicting drug targets and mechanisms of action
bioRxiv. (2019). DOI: 10.1101/580654
Moreno P,
Pireddu L,
Roger P,
Goonasekera N,
Afgan E,
Beek Mvd,
He S,
Larsson A,
Ruttkies C,
Schober D,
Johnson D,
Rocca-Serra P,
Weber RJM,
Gruening B,
Salek B,
Kale N,
Perez-Riverol Y,
Papatheodorou I,
Spjuth O,
Neumann D
Galaxy-Kubernetes integration: scaling bioinformatics workflows in the cloud
bioRxiv. 488643 (2018). DOI: 10.1101/488643
Galaxy-Kubernetes integration: scaling bioinformatics workflows in the cloud
bioRxiv. 488643 (2018). DOI: 10.1101/488643
Spjuth O,
Carlsson L.,
Gauraha N.
Aggregating Predictions on Multiple Non-disclosed Datasets using Conformal Prediction
arXiv. 1806.04000 (2018). URL: arxiv.org/abs/1806.04000
Aggregating Predictions on Multiple Non-disclosed Datasets using Conformal Prediction
arXiv. 1806.04000 (2018). URL: arxiv.org/abs/1806.04000
Gauraha N,
Spjuth O.
conformalClassification: A Conformal Prediction R Package for Classification.
arXiv:1804.05494. (2018). URL: arxiv.org/abs/1804.05494
conformalClassification: A Conformal Prediction R Package for Classification.
arXiv:1804.05494. (2018). URL: arxiv.org/abs/1804.05494
2025
Alejandro Lagunas-Rangel F,
Åberg M,
Liao S,
Espinelli-Amorim F,
Tummaramatti-Hanumant R,
Jackeviča L,
Briviba M,
Liu W,
Alsehli A,
Fredriksson R,
Spjuth O,
Klovins J,
Dambrova M,
Kudłak B,
Andersson C,
and Schiöth H.
Extensive analysis of pollutant interactions: Identification of deleterious synergistic effects at environmentally relevant dose levels using Drosophila and mammalian cells
Science of the Total Environment. 992, 179848 (2025). DOI: 10.1016/j.scitotenv.2025.179848
Extensive analysis of pollutant interactions: Identification of deleterious synergistic effects at environmentally relevant dose levels using Drosophila and mammalian cells
Science of the Total Environment. 992, 179848 (2025). DOI: 10.1016/j.scitotenv.2025.179848
Seal S,
Mahale M,
Garcia-Ortegon M,
Joshi C,
Hosseini-Gerami L,
Beatson A,
Greenig M,
Shekhar M,
Patra A,
Weis C,
Mehrjou A,
Badré A,
Paisley B,
Lowe R,
Singh S,
Shah F,
Johannesson B,
Williams D,
Rouquié D,
Clevert DA,
Schwab P,
Richmond N,
Nicolaou C,
Gonzalez R,
Naven R,
Schramm C,
Vidler L,
Mansouri K,
Walters WP,
Dalmas Wilk D,
Spjuth O,
Carpenter AE,
and Bender A.
Machine Learning for Toxicity Prediction Using Chemical Structure: Pillars for Success in the Real World
Chemical Research in Toxicology. (2025). DOI: 10.1021/acs.chemrestox.5c00033
Machine Learning for Toxicity Prediction Using Chemical Structure: Pillars for Success in the Real World
Chemical Research in Toxicology. (2025). DOI: 10.1021/acs.chemrestox.5c00033
Sreenivasan AP,
Vaivade A,
Noui Y,
Khoonsari PE,
Burman J,
Spjuth O,
Kultima K.
Conformal prediction enables disease course prediction and allows individualized diagnostic uncertainty in multiple sclerosis
npj Digital Medicine. 8, 224 (2025). DOI: 10.1038/s41746-025-01616-z
Conformal prediction enables disease course prediction and allows individualized diagnostic uncertainty in multiple sclerosis
npj Digital Medicine. 8, 224 (2025). DOI: 10.1038/s41746-025-01616-z
Tanoli Z,
Fernández-Torras A,
Özcan UO,
Kushnir A,
Michelle Nader K,
Gadiya Y,
Fiorenza L,
Ianevski A,
Vähä-Koskela M,
Miihkinen M,
Seemab U,
LeinonenH,
Seashore-Ludlow B,
Tampere M,
Kalman A,
Ballante F,
Benfenati E,
Saunders G,
Potdar S,
Gómez García I,
García-Serna R,
Talarico C,
Rosario Beccari A,
Schaal W,
Polo A,
Costantini S,
Cabri E,
Jacobs M,
Saarela J,
Budillon A,
Spjuth O,
Östling P,
Xhaard H,
Quintana J,
Mestres J,
Gribbon P,
Ussi AE,
Lo D,
de Kort M,
Wennerberg K,
Fratelli M,
Carreras-Puigvert J and Aittokallio T
Computational drug repurposing: approaches, evaluation of in silico resources and case studies
Nature Reviews Drug Discovery. (2025). DOI: 10.1038/s41573-025-01164-x
Computational drug repurposing: approaches, evaluation of in silico resources and case studies
Nature Reviews Drug Discovery. (2025). DOI: 10.1038/s41573-025-01164-x
Vaivade A,
Erngren I,
Carlsson G,
Freyhult E,
Khoonsari PE,
Noui Y,
Al-Grety A,
Åkerfeldt T,
Spjuth O,
Gallo V,
Kockum I,
Alfredsson L,
Olsson T,
Burman J,
and Kultima K
Associations of PFAS and OH-PCBs with Risk of Multiple Sclerosis Onset and Disability Worsening
Nature Communications. 16, 2014 (2025). DOI: 10.1038/s41467-025-57172-3
Associations of PFAS and OH-PCBs with Risk of Multiple Sclerosis Onset and Disability Worsening
Nature Communications. 16, 2014 (2025). DOI: 10.1038/s41467-025-57172-3
Gindullin R,
Francisco Rodríguez MA,
Seashore-Ludlow B,
and Spjuth O.
Minimising Source-Plate Swaps for Robotised Compound Dispensing in Microplates
22nd International Conference on the Integration of Constraint Programming, Artificial Intelligence, and Operations Research (CPAIOR 2025). Accepted (2025). URL: openreview.net/forum?id=zwfRw4zCEm
Minimising Source-Plate Swaps for Robotised Compound Dispensing in Microplates
22nd International Conference on the Integration of Constraint Programming, Artificial Intelligence, and Operations Research (CPAIOR 2025). Accepted (2025). URL: openreview.net/forum?id=zwfRw4zCEm
Seal S,
Trapotsi MA,
Spjuth O,
Singh S,
Carreras-Puigvert J,
Greene N,
Bender A,
Carpenter AE
Cell Painting: A Decade of Discovery and Innovation in Cellular Imaging
Nature Methods. 22, 254-268. (2025). DOI: 10.1038/s41592-024-02528-8
Cell Painting: A Decade of Discovery and Innovation in Cellular Imaging
Nature Methods. 22, 254-268. (2025). DOI: 10.1038/s41592-024-02528-8
2024
Seal S,
Williams DP,
Hosseini-Gerami L,
Mahael M,
Carpenter AE,
Spjuth O,
and Bender A.
Improved Early Detection of Drug-Induced Liver Injury by Integrating Predicted in vivo and in vitro Data
Chemical Research in Toxicology. 37, 8, 1290–1305. (2024). DOI: 10.1021/acs.chemrestox.4c00015
Improved Early Detection of Drug-Induced Liver Injury by Integrating Predicted in vivo and in vitro Data
Chemical Research in Toxicology. 37, 8, 1290–1305. (2024). DOI: 10.1021/acs.chemrestox.4c00015
Porebski B,
Christ W,
Corman A,
Haraldsson M,
Barz M,
Lidemalm L,
Häggblad M,
Ilmain J,
Wright SC,
Murga M,
Schlegel J,
Jarvius M,
Lapins M,
Sezgin E,
Bhabha G,
Lauschke VM,
Jordi Carreras-Puigvert J,
Lafarga M,
Klingström J,
Hühn D,
Fernandez-Capetillo O.
Discovery of a novel inhibitor of macropinocytosis with antiviral activity. Molecular Therapy
Molecular Therapy. 32, 9, 3012-3024. (2024). DOI: 10.1016/j.ymthe.2024.06.038
Discovery of a novel inhibitor of macropinocytosis with antiviral activity. Molecular Therapy
Molecular Therapy. 32, 9, 3012-3024. (2024). DOI: 10.1016/j.ymthe.2024.06.038
Arvidsson McShane S,
Norinder U,
Alvarsson J,
Ahlberg E,
Carlsson L,
and Spjuth O.
CPSign - Conformal Prediction for Cheminformatics Modeling
Journal of Cheminformatics. 16, 75 (2024). DOI: 10.1186/s13321-024-00870-9
CPSign - Conformal Prediction for Cheminformatics Modeling
Journal of Cheminformatics. 16, 75 (2024). DOI: 10.1186/s13321-024-00870-9
Ju L,
Hellander A,
and Spjuth O.
Federated Learning for Predicting Compound Mechanism of Action Based on Image-data from Cell Painting
Artificial Intelligence in Life Sciences. 5, 100098 (2024). DOI: 10.1016/j.ailsci.2024.100098
Federated Learning for Predicting Compound Mechanism of Action Based on Image-data from Cell Painting
Artificial Intelligence in Life Sciences. 5, 100098 (2024). DOI: 10.1016/j.ailsci.2024.100098
Carreras-Puigvert J,
and Spjuth O
Artificial Intelligence for High Content Imaging in Drug Discovery
Current Opinion in Structural Biology. 87, 102842 (2024). DOI: 10.1016/j.sbi.2024.102842
Artificial Intelligence for High Content Imaging in Drug Discovery
Current Opinion in Structural Biology. 87, 102842 (2024). DOI: 10.1016/j.sbi.2024.102842
Tal T,
Myhre O,
Fritsche E,
Rüegg J,
Craenen K,
Aiello K,
Agrillo C,
Babin PJ,
Escher BI,
Dirven H,
Hellsten K,
Dolva K,
Heusinkveld H,
Hadzhiev Y,
Hurem S,
Jagiello K,
Judzinska B,
Klüver N,
Knoll-Gellida A,
Kühne BA,
Leist M,
Lislien M,
Lyche JL,
Müller F,
Neuhaus W,
Pallocca G,
Seeger B,
Scharkin I,
Scholz S,
Spjuth O,
Torres-Ruiz M and Bartmann K.
New approach methods to assess developmental and adult neurotoxicity for regulatory use: A PARC Work Package 5 project
Frontiers in Toxicology. 6, 1359507 (2024). DOI: 10.3389/ftox.2024.1359507
New approach methods to assess developmental and adult neurotoxicity for regulatory use: A PARC Work Package 5 project
Frontiers in Toxicology. 6, 1359507 (2024). DOI: 10.3389/ftox.2024.1359507
Zhang T,
Gupta A,
Rodríguez MAF,
Spjuth O,
Hellander A and Toor S.
Data management of scientific applications in a reinforcement learning-based hierarchical storage system
Expert Systems With Applications. 237, 121443 (2024). DOI: 10.1016/j.eswa.2023.121443
Data management of scientific applications in a reinforcement learning-based hierarchical storage system
Expert Systems With Applications. 237, 121443 (2024). DOI: 10.1016/j.eswa.2023.121443
Seal S,
Carreras-Puigvert J,
Carpenter AE,
Spjuth O,
Bender A.
From Pixels to Phenotypes: Integrating Image-Based Profiling with Cell Health Data Improves Interpretability
Molecular Biology of the Cell. 35, 3 (2024). DOI: 10.1091/mbc.E23-08-0298
From Pixels to Phenotypes: Integrating Image-Based Profiling with Cell Health Data Improves Interpretability
Molecular Biology of the Cell. 35, 3 (2024). DOI: 10.1091/mbc.E23-08-0298
Seal S,
Spjuth O,
Hosseini-Gerami L,
Garcia-Ortegon M,
Singh S,
Bender A,
Carpenter AE.
Insights into Drug Cardiotoxicity from Biological and Chemical Data: The First Public Classifiers for FDA DICTrank
Journal of Chemical Information and Modeling. 64, 4, 1172-1186. (2024). DOI: 10.1021/acs.jcim.3c01834
Insights into Drug Cardiotoxicity from Biological and Chemical Data: The First Public Classifiers for FDA DICTrank
Journal of Chemical Information and Modeling. 64, 4, 1172-1186. (2024). DOI: 10.1021/acs.jcim.3c01834
2023
Harrison PJ,
Gupta A,
Rietdijk J,
Wieslander H,
Carreras-Puigvert J,
Georgiev P,
Wählby C,
Spjuth O,
Sintorn IM.
Evaluating the utility of brightfield image data for mechanism of action prediction
PLOS Computational Biology. 19, 7, e1011323. (2023). DOI: 10.1371/journal.pcbi.1011323
Evaluating the utility of brightfield image data for mechanism of action prediction
PLOS Computational Biology. 19, 7, e1011323. (2023). DOI: 10.1371/journal.pcbi.1011323
Braeuning A,
Balaguer P,
Bourguet W,
Carreras-Puigvert J,
Feiertag K,
Kamstra JH,
Knapen D,
Lichtenstein D,
Marx-Stoelting P,
Rietdijk J,
Schubert K,
Spjuth O,
Stinckens E,
Thedieck K,
van den Boom R,
Vergauwen L,
Von Bergen M,
Wewer N and Zalko D.
Development of new approach methods for the identification and characterization of endocrine metabolic disruptors – a PARC project
Frontiers in Toxicology. 5, 1212509 (2023). DOI: 10.3389/ftox.2023.1212509
Development of new approach methods for the identification and characterization of endocrine metabolic disruptors – a PARC project
Frontiers in Toxicology. 5, 1212509 (2023). DOI: 10.3389/ftox.2023.1212509
Herman S,
Arvidsson McShane S,
Zhukovsky C,
Khoonsari PE,
Svenningsson A,
Burman J,
Spjuth O,
Kultima K.
Disease phenotype prediction in multiple sclerosis
iScience. 26, 6 (2023). DOI: 10.1016/j.isci.2023.106906
Disease phenotype prediction in multiple sclerosis
iScience. 26, 6 (2023). DOI: 10.1016/j.isci.2023.106906
Seal S,
Yang H,
Trapotsi MA,
Singh S,
Carreras-Puigvert J,
Spjuth O,
Bender A
Merging Bioactivity Predictions from Cell Morphology and Chemical Fingerprint Models by Leveraging Similarity to Training Data
Journal of Cheminformatics. 15, 56 (2023). DOI: 10.1186/s13321-023-00723-x
Merging Bioactivity Predictions from Cell Morphology and Chemical Fingerprint Models by Leveraging Similarity to Training Data
Journal of Cheminformatics. 15, 56 (2023). DOI: 10.1186/s13321-023-00723-x
Rodríguez MAF,
Carreras-Puigvert J,
and Spjuth O
Designing microplate layouts using artificial intelligence
Artificial Intelligence in the Life Sciences. 3, 100073 (2023). DOI: 10.1016/j.ailsci.2023.100073
Designing microplate layouts using artificial intelligence
Artificial Intelligence in the Life Sciences. 3, 100073 (2023). DOI: 10.1016/j.ailsci.2023.100073
Tian G,
Harrison PJ,
Sreenivasan AP,
Carreras-Puigvert J,
Spjuth O
Combining molecular and cell painting image data for mechanism of action prediction
Artificial Intelligence in Life Science. 3, 100060 (2023). DOI: 10.1016/j.ailsci.2023.100060
Combining molecular and cell painting image data for mechanism of action prediction
Artificial Intelligence in Life Science. 3, 100060 (2023). DOI: 10.1016/j.ailsci.2023.100060
Fagerholm U,
Hellberg S,
Alvarsson J and Spjuth O.
In silico prediction of human clinical pharmacokinetics with ANDROMEDA by Prosilico – Predictions for a proposed benchmarking data set and new small drugs on the market 2021 and comparison with laboratory methods
Alternatives to Laboratory Animals. 51, 1, 39-54. (2023). DOI: 10.1177/02611929221148447
In silico prediction of human clinical pharmacokinetics with ANDROMEDA by Prosilico – Predictions for a proposed benchmarking data set and new small drugs on the market 2021 and comparison with laboratory methods
Alternatives to Laboratory Animals. 51, 1, 39-54. (2023). DOI: 10.1177/02611929221148447
2022
Olsson H,
Kartasalo K,
Mulliqi N,
Capuccini M,
Ruusuvuori P,
Samaratunga H,
Delahunt B,
Lindskog C,
Janssen E,
Billie A,
Egevad L,
Spjuth O,
and Eklund M.
Estimating diagnostic uncertainty in artificial intelligence assisted pathology using conformal prediction
Nature Communications. 13, 7761 (2022). DOI: 10.1038/s41467-022-34945-8
Estimating diagnostic uncertainty in artificial intelligence assisted pathology using conformal prediction
Nature Communications. 13, 7761 (2022). DOI: 10.1038/s41467-022-34945-8
Sheffield N,
Bonazzi V,
Bourne P,
Burdett T,
Clark T,
Grossman R,
Spjuth O and Yates A.
From biomedical cloud platforms to microservices: next steps in FAIR data and analysis
Nature Scientific Data. 9, 553 (2022). DOI: 10.1038/s41597-022-01619-5
From biomedical cloud platforms to microservices: next steps in FAIR data and analysis
Nature Scientific Data. 9, 553 (2022). DOI: 10.1038/s41597-022-01619-5
Fagerholm U,
Spjuth O,
and Hellberg S.
The impact of reference data selection for the prediction accuracy of intrinsic hepatic metabolic clearance
Journal of Pharmaceutical Sciences. 111, 9, 2645-2649. (2022). DOI: 10.1016/j.xphs.2022.06.024
The impact of reference data selection for the prediction accuracy of intrinsic hepatic metabolic clearance
Journal of Pharmaceutical Sciences. 111, 9, 2645-2649. (2022). DOI: 10.1016/j.xphs.2022.06.024
Seal S,
Carreras-Puigvert J,
Trapotsi MA,
Yang H,
Spjuth O,
Bender A
Integrating Cell Morphology with Gene Expression and Chemical Structure to Aid Mitochondrial Toxicity Detection
Nature Communications Biology. 5, 858 (2022). DOI: 10.1038/s42003-022-03763-5
Integrating Cell Morphology with Gene Expression and Chemical Structure to Aid Mitochondrial Toxicity Detection
Nature Communications Biology. 5, 858 (2022). DOI: 10.1038/s42003-022-03763-5
Raykova D,
Kermpatsou D,
Malmqvist T,
Harrison P,
Sander MR,
Stiller C,
Heldin J,
Leino M,
Ricardo S,
Klemm A,
David L,
Spjuth O,
Vemuri K,
Dimberg A,
Sundqvist A,
Norlin M,
Klaesson A,
Kampf C and Söderberg O.
A method for Boolean analysis of protein interactions at a molecular level
Nature Communications. 13, 4755 (2022). DOI: 10.1038/s41467-022-32395-w
A method for Boolean analysis of protein interactions at a molecular level
Nature Communications. 13, 4755 (2022). DOI: 10.1038/s41467-022-32395-w
Lukashina N,
Kartysheva E,
Spjuth O,
Virko E and Shpilman A.
SimVec: predicting polypharmacy side effects for new drugs
Journal of Cheminformatics. 14, 49 (2022). DOI: 10.1186/s13321-022-00632-5
SimVec: predicting polypharmacy side effects for new drugs
Journal of Cheminformatics. 14, 49 (2022). DOI: 10.1186/s13321-022-00632-5
Schaal W,
Ameur A,
Olsson-Strömberg U,
Hermanson M,
Cavelier L,
and Spjuth O
Migrating to Long-Read Sequencing for Clinical Routine BCR-ABL1 TKI Resistance Mutation Screening
Cancer Informatics. 21, 1-8. (2022). DOI: 10.1177/11769351221110872
Migrating to Long-Read Sequencing for Clinical Routine BCR-ABL1 TKI Resistance Mutation Screening
Cancer Informatics. 21, 1-8. (2022). DOI: 10.1177/11769351221110872
Sreenivasan AP,
Harrison PJ,
Schaal W,
Matuszewski DJ,
Kultima K,
and Spjuth O.
Predicting protein network topology clusters from chemical structure using deep learning
Journal of Cheminformatics. 14, 47 (2022). DOI: 10.1186/s13321-022-00622-7
Predicting protein network topology clusters from chemical structure using deep learning
Journal of Cheminformatics. 14, 47 (2022). DOI: 10.1186/s13321-022-00622-7
Ekmefjord M,
Ait-Mlouk A,
Alawadi S,
Åkesson M,
Stoyanova D,
Spjuth O,
Toor S,
Hellander A
Scalable federated machine learning with FEDn
The 22nd IEEE/ACM International Symposium on Cluster, Cloud and Internet Computing (CCGrid). , 555-564. (2022). DOI: 10.1109/CCGrid54584.2022.00065
Scalable federated machine learning with FEDn
The 22nd IEEE/ACM International Symposium on Cluster, Cloud and Internet Computing (CCGrid). , 555-564. (2022). DOI: 10.1109/CCGrid54584.2022.00065
Fagerholm U,
Hellberg S,
Alvarsson J,
and Spjuth O.
In silico predictions of the gastrointestinal uptake of macrocycles in man using conformal prediction methodology
Journal of Pharmaceutical Sciences. 111, 9, 2614-2619. (2022). DOI: 10.1016/j.xphs.2022.05.010
In silico predictions of the gastrointestinal uptake of macrocycles in man using conformal prediction methodology
Journal of Pharmaceutical Sciences. 111, 9, 2614-2619. (2022). DOI: 10.1016/j.xphs.2022.05.010
Rietdijk J,
Aggarwal T,
Georgieva P,
Lapins M,
Carreras-Puigvert J,
and Spjuth O.
Morphological profiling of environmental chemicals enables efficient and untargeted exploration of combination effects
Science of the Total Environment. 832, 155058 (2022). DOI: 10.1016/j.scitotenv.2022.155058
Morphological profiling of environmental chemicals enables efficient and untargeted exploration of combination effects
Science of the Total Environment. 832, 155058 (2022). DOI: 10.1016/j.scitotenv.2022.155058
Fagerholm U,
Hellberg S,
Alvarsson J,
and Spjuth O.
In silico predictions of the human pharmacokinetics/toxicokinetics of 65 chemicals from various classes using conformal prediction methodology
Xenobiotica. 52, 2, 113-118. (2022). DOI: 10.1080/00498254.2022.2049397
In silico predictions of the human pharmacokinetics/toxicokinetics of 65 chemicals from various classes using conformal prediction methodology
Xenobiotica. 52, 2, 113-118. (2022). DOI: 10.1080/00498254.2022.2049397
2021
Fagerholm U,
Hellberg S,
Alvarsson A,
Arvidsson McShane S,
and Spjuth O.
In silico predictions of volume of distribution of drugs in man using conformal prediction performs on par with animal data-based models
Xenobiotica. 51, 12, 1366-1371. (2021). DOI: 10.1080/00498254.2021.2011471
In silico predictions of volume of distribution of drugs in man using conformal prediction performs on par with animal data-based models
Xenobiotica. 51, 12, 1366-1371. (2021). DOI: 10.1080/00498254.2021.2011471
Martens M,
Stierum R,
Schymanski EL,
Evelo CT,
Aalizadeh R,
Aladjov H,
Arturi K,
Audouze K,
Babica P,
Berka K,
Bessems J,
Blaha L,
Bolton EE,
Cases M,
Damalas D,
Dave K,
Dilger M,
Exner T,
Geerke DP,
Grafstrom R,
Gray A,
Hancock JM,
Hollert H,
Jeliazkova N,
Jennen D,
Jourdan F,
Kahlem P,
Klanova J,
Kleinjans J,
Kondic T,
Kone B,
Lynch I,
Maran U,
Martinez Cuesta S,
Menager H,
Neumann S,
Nymark P,
Oberacher H,
Ramirez N,
Remy S,
Rocca-Serra P,
Salek RM,
Sallach B,
Sansone SA,
Sanz F,
Sarimveis H,
Sarntivijai S,
Schulze T,
Slobodnik J,
Spjuth O,
Tedds J,
Thomaidis N,
Weber RJM,
van Westen GJP,
Wheelock CE,
Williams AJ,
Witters H,
Zdrazil B,
Zupanic A,
Willighagen EL.
ELIXIR and Toxicology: a community in development
F1000Research. 10(ELIXIR), 1129 (2021). DOI: 10.12688/f1000research.74502.1
ELIXIR and Toxicology: a community in development
F1000Research. 10(ELIXIR), 1129 (2021). DOI: 10.12688/f1000research.74502.1
Wieslander H,
Gupta A,
Bergman E,
Hallström E,
Harrison PJ
Learning to see colours: generating biologically relevant fluorescent labels from bright-field images
PLOS ONE. 16, 10 (2021). DOI: 10.1371/journal.pone.0258546
Learning to see colours: generating biologically relevant fluorescent labels from bright-field images
PLOS ONE. 16, 10 (2021). DOI: 10.1371/journal.pone.0258546
Norinder U,
Spjuth O,
Svensson F.
Synergy conformal prediction applied to large-scale bioactivity datasets and in federated learning
Journal of Cheminformatics. 13, 77 (2021). DOI: 10.1186/s13321-021-00555-7
Synergy conformal prediction applied to large-scale bioactivity datasets and in federated learning
Journal of Cheminformatics. 13, 77 (2021). DOI: 10.1186/s13321-021-00555-7
Lukashina N,
Williams MJ,
Kartysheva E,
Virko E,
Kudłak B,
Fredriksson R,
Spjuth O,
Schiöth HB.
Integrating statistical and machine-learning approach for meta-analysis of Bisphenol A-exposure datasets reveals effects on mouse gene expression within pathways of apoptosis and cell survival
International Journal of Molecular Sciences. 22, 19 (2021). DOI: 10.3390/ijms221910785
Integrating statistical and machine-learning approach for meta-analysis of Bisphenol A-exposure datasets reveals effects on mouse gene expression within pathways of apoptosis and cell survival
International Journal of Molecular Sciences. 22, 19 (2021). DOI: 10.3390/ijms221910785
Ouyang W,
Bowman R,
Wang H,
Bumke KE,
Collins JT,
Spjuth O,
Carreras-Puigvert J and Diederich B
An Open-Source Modular Framework for Automated Pipetting and Imaging Applications
Advanced Biology. 2101063 (2021). DOI: 10.1002/adbi.202101063
An Open-Source Modular Framework for Automated Pipetting and Imaging Applications
Advanced Biology. 2101063 (2021). DOI: 10.1002/adbi.202101063
Fagerholm U,
Spjuth O,
and Hellberg S.
Comparison between Lab Variability and in silico Prediction Errors for the Unbound Fraction of Drugs in Human Plasma
Xenobiotica. 51, 10, 1095-1100. (2021). DOI: 10.1080/00498254.2021.1964044
Comparison between Lab Variability and in silico Prediction Errors for the Unbound Fraction of Drugs in Human Plasma
Xenobiotica. 51, 10, 1095-1100. (2021). DOI: 10.1080/00498254.2021.1964044
Rietdijk J,
Tampere M,
Pettke A,
Georgieva P,
Lapins M,
Warpman Berglund U,
Spjuth O,
Puumalainen MR,
Carreras-Puigvert J
A phenomics approach for antiviral drug discovery
BMC Biology. 19, 156 (2021). DOI: 10.1186/s12915-021-01086-1
A phenomics approach for antiviral drug discovery
BMC Biology. 19, 156 (2021). DOI: 10.1186/s12915-021-01086-1
Gauraha,
N. and Spjuth,
O.
Synergy Conformal Prediction
Proceedings of the Tenth Symposium on Conformal and Probabilistic Prediction and Applications, PMLR. 152, 91-110. (2021). URL: proceedings.mlr.press/v152/gauraha21a.html
Synergy Conformal Prediction
Proceedings of the Tenth Symposium on Conformal and Probabilistic Prediction and Applications, PMLR. 152, 91-110. (2021). URL: proceedings.mlr.press/v152/gauraha21a.html
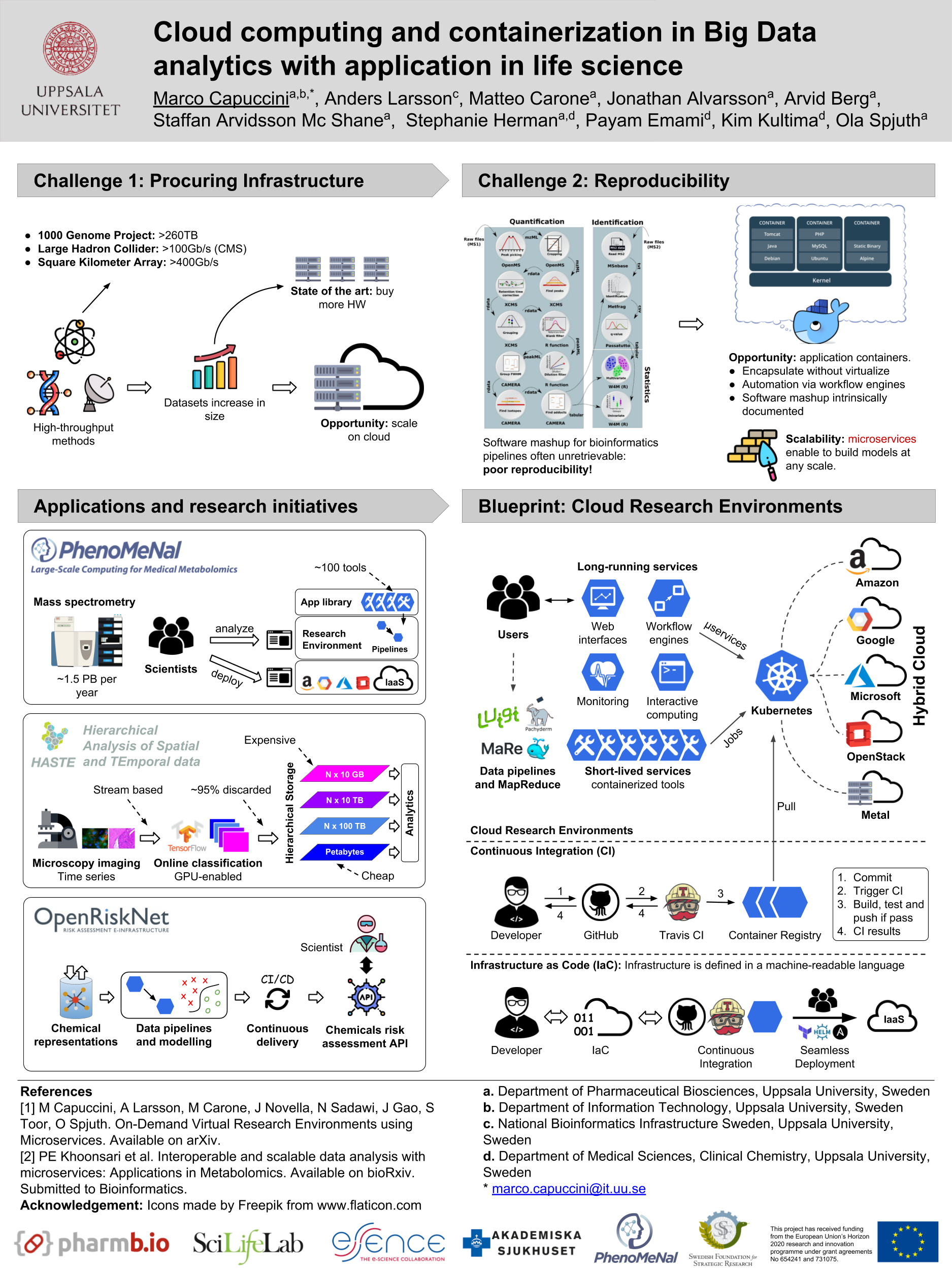
Spjuth O,
Capuccini M,
Carone M,
Larsson A,
Schaal W,
Novella J,
Stein JA,
Ekmefjord M,
Di Tommaso P,
Floden E,
Notredame C,
Moreno P,
Khoonsari PE,
Herman S,
Kultima K,
Lampa S
Approaches for containerized scientific workflows in cloud environments with applications in life science
F1000Research. 10, 513 (2021). DOI: 10.12688/f1000research.53698.1
Approaches for containerized scientific workflows in cloud environments with applications in life science
F1000Research. 10, 513 (2021). DOI: 10.12688/f1000research.53698.1
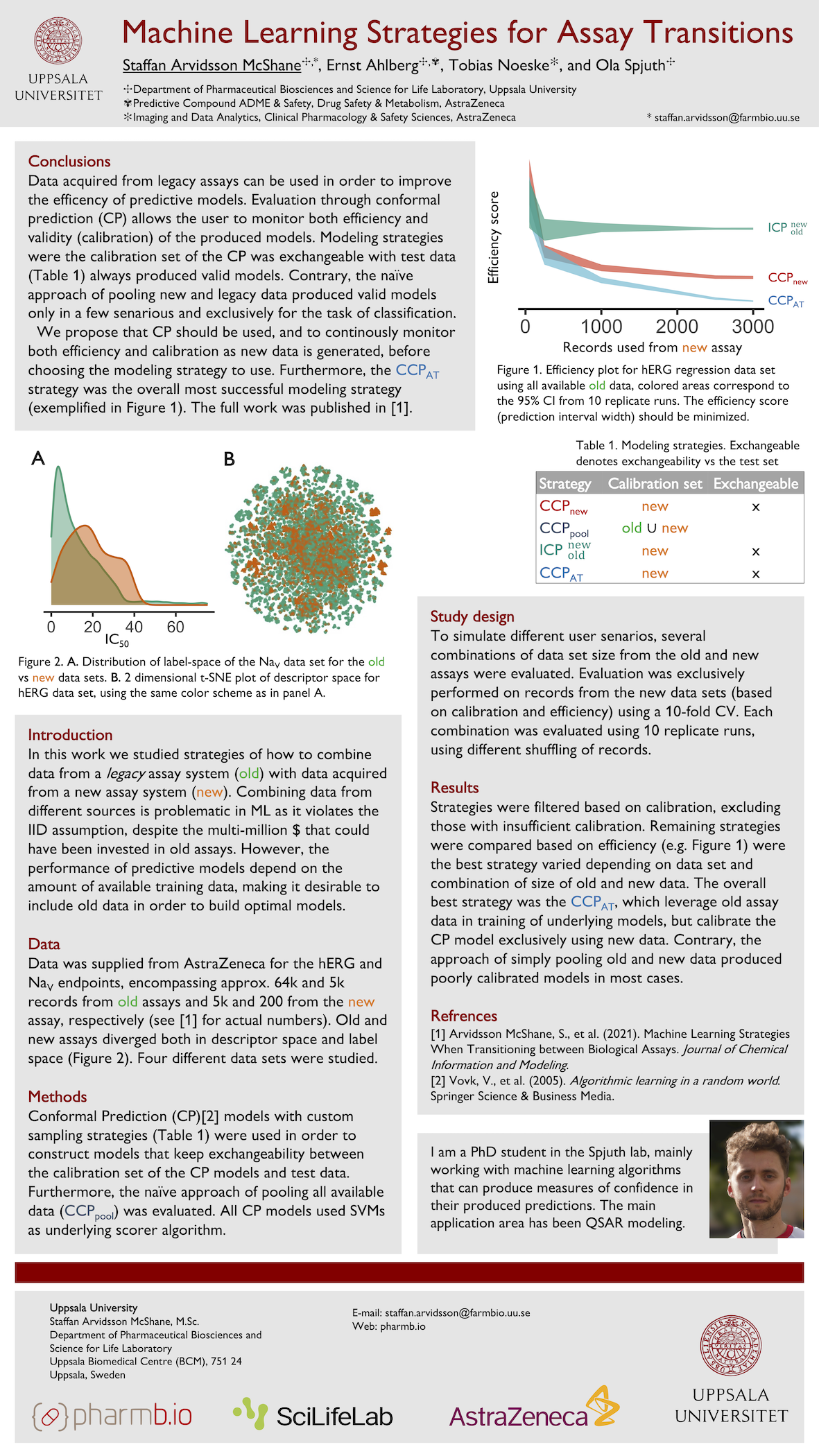
Arvidsson McShane S,
Ahlberg E,
Noeske T,
and Spjuth O.
Machine learning strategies when transitioning between biological assays
Journal of Chemical Information and Modeling. 61, 7, 3722-3733. (2021). DOI: 10.1021/acs.jcim.1c00293
Machine learning strategies when transitioning between biological assays
Journal of Chemical Information and Modeling. 61, 7, 3722-3733. (2021). DOI: 10.1021/acs.jcim.1c00293
Spjuth O,
Frid J,
and Hellander A.
The Machine Learning Life Cycle and the Cloud: Implications for Drug Discovery
Expert Opinion On Drug Discovery. 16, 9, 1071-1079. (2021). DOI: 10.1080/17460441.2021.1932812
The Machine Learning Life Cycle and the Cloud: Implications for Drug Discovery
Expert Opinion On Drug Discovery. 16, 9, 1071-1079. (2021). DOI: 10.1080/17460441.2021.1932812
Fagerholm U,
Hellberg S,
and Spjuth O.
Advances in Predictions of Oral Bioavailability of Candidate Drugs in Man with New Machine Learning Methodology
Molecules. 6, 29 (2021). DOI: 10.3390/molecules26092572
Advances in Predictions of Oral Bioavailability of Candidate Drugs in Man with New Machine Learning Methodology
Molecules. 6, 29 (2021). DOI: 10.3390/molecules26092572
Morger A,
Svensson F,
Arvidsson McShane S,
Gauraha N,
Norinder U,
Spjuth O,
Volkamer A
Assessing the Calibration in Toxicological in Vitro Models with Conformal Prediction
Journal of Cheminformatics. 13, 35 (2021). DOI: 10.1186/s13321-021-00511-5
Assessing the Calibration in Toxicological in Vitro Models with Conformal Prediction
Journal of Cheminformatics. 13, 35 (2021). DOI: 10.1186/s13321-021-00511-5
Jakobsson J,
Spjuth O,
Lagerström M.
scConnect: a method for exploratory analysis of cell-cell communication based on single cell RNA sequencing data
Bioinformatics. 37, 20, 3501–3508. (2021). DOI: 10.1093/bioinformatics/btab245
scConnect: a method for exploratory analysis of cell-cell communication based on single cell RNA sequencing data
Bioinformatics. 37, 20, 3501–3508. (2021). DOI: 10.1093/bioinformatics/btab245
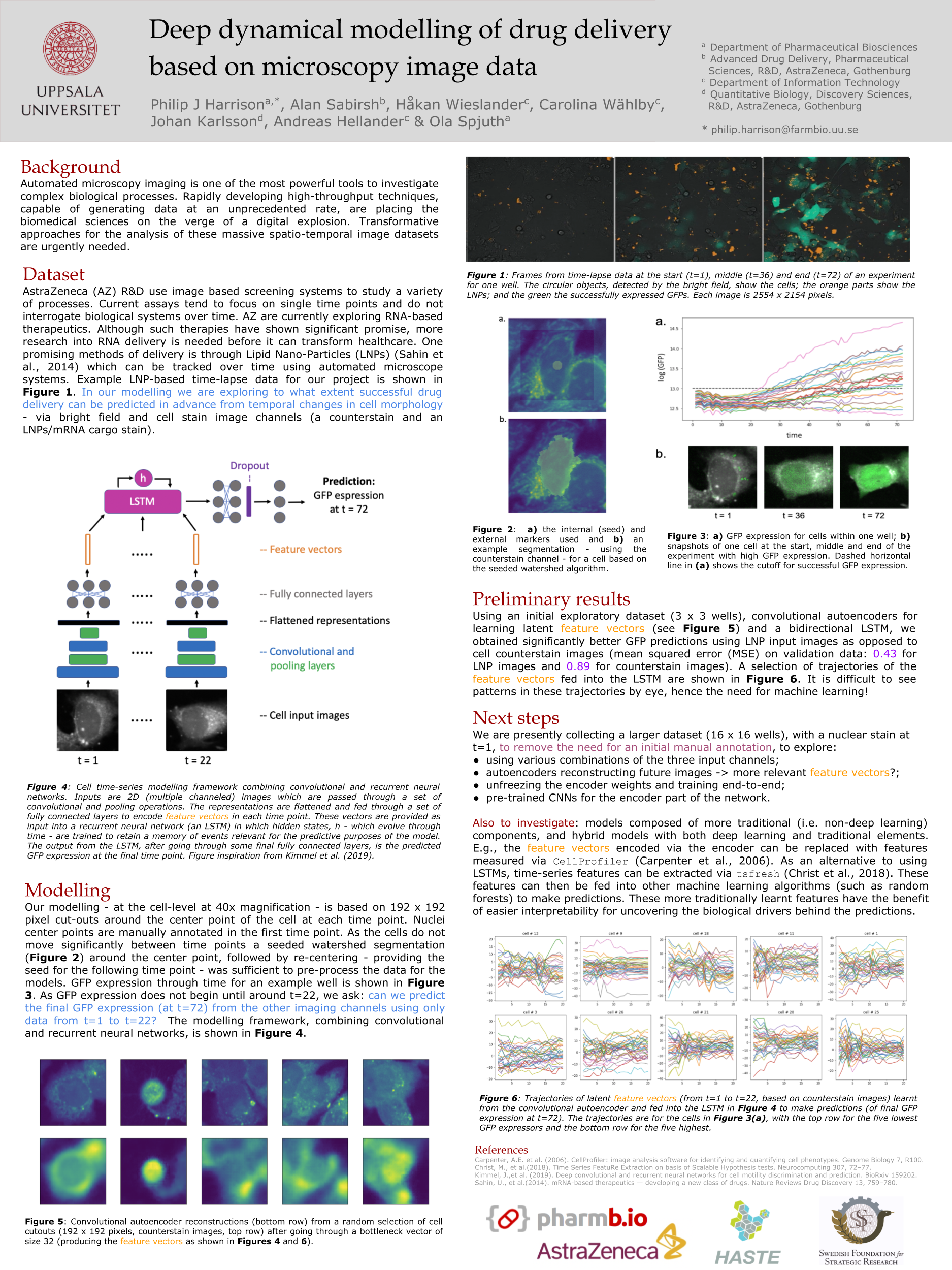
Harrison P,
Wieslander H,
Sabirsh A,
Karlsson J,
Malmsjö V,
Hellander A,
Wählby C,
Spjuth O
Deep learning models for lipid-nanoparticle-based drug delivery
Nanomedicine. 16, 13 (2021). DOI: 10.2217/nnm-2020-0461
Deep learning models for lipid-nanoparticle-based drug delivery
Nanomedicine. 16, 13 (2021). DOI: 10.2217/nnm-2020-0461
Blamey B,
Toor S,
Dahlö M,
Wieslander H,
Harrison PJ,
Sintorn IM,
Sabirsh A,
Wählby C,
Spjuth O,
Hellander A
Rapid development of cloud-native intelligent data pipelines for scientific data streams using the HASTE Toolkit
Gigascience. 10, 3 (2021). DOI: 10.1093/gigascience/giab018
Rapid development of cloud-native intelligent data pipelines for scientific data streams using the HASTE Toolkit
Gigascience. 10, 3 (2021). DOI: 10.1093/gigascience/giab018
Gauraha,
N. and Spjuth,
O.
Synergy Conformal Prediction for Regression
Proceedings of the 10th International Conference on Pattern Recognition Applications and Methods. vol 1: ICPRAM, 212-221. (2021). DOI: 10.5220/0010229402120221
Synergy Conformal Prediction for Regression
Proceedings of the 10th International Conference on Pattern Recognition Applications and Methods. vol 1: ICPRAM, 212-221. (2021). DOI: 10.5220/0010229402120221
Alvarsson J,
Arvidsson McShane S,
Norinder U and Spjuth O.
Predicting with confidence: Using conformal prediction in drug discovery
Journal of Pharmaceutical Sciences. 110, 1, 42-49. (2021). DOI: 10.1016/j.xphs.2020.09.055
Predicting with confidence: Using conformal prediction in drug discovery
Journal of Pharmaceutical Sciences. 110, 1, 42-49. (2021). DOI: 10.1016/j.xphs.2020.09.055
Wieslander H.,
Harrison P,
Skogberg G,
Jackson S,
Fridén M,
Karlsson J,
Spjuth O,
and Wählby C.
Deep learning with conformal prediction for hierarchical analysis of large-scale whole-slide tissue images
IEEE Journal of Biomedical and Health Informatics. 25, 2, 371-380. (2021). DOI: 10.1109/JBHI.2020.2996300
Deep learning with conformal prediction for hierarchical analysis of large-scale whole-slide tissue images
IEEE Journal of Biomedical and Health Informatics. 25, 2, 371-380. (2021). DOI: 10.1109/JBHI.2020.2996300
2020
Ashrafian H,
Glen R,
Ebbels T,
Blaise B,
Kalra D,
Kultima K,
Spjuth O,
Tenori L,
Sounderajah V,
Salek RM,
Kale N,
Haug K,
Schober D,
Rocca-Serra P,
O'Donovan C,
Steinbeck C,
Cano I,
de Atauri P,
Cascante M
Metabolomics - the stethoscope for the 21st century
Medical Principles and Practice. 30, 301–310. (2020). DOI: 10.1159/000513545
Metabolomics - the stethoscope for the 21st century
Medical Principles and Practice. 30, 301–310. (2020). DOI: 10.1159/000513545
Stein O,
Blamey B,
Karlsson J,
Sabirsh A,
Spjuth O,
Hellander A,
and Toor S.
Smart Resource Management for Data Streaming using an Online Bin-packing Strategy
2020 IEEE International Conference on Big Data. , 2207-2216. (2020). DOI: 10.1109/BigData50022.2020.9378241
Smart Resource Management for Data Streaming using an Online Bin-packing Strategy
2020 IEEE International Conference on Big Data. , 2207-2216. (2020). DOI: 10.1109/BigData50022.2020.9378241
Ahmed L,
Alogheli H,
Arvidsson McShane S,
Alvarsson J,
Berg A,
Larsson A,
Schaal W,
Laure E and Spjuth O.
Predicting Target Profiles with Confidence as a Service using Docking Scores
Journal of Cheminformatics. 12, 62 (2020). DOI: 10.1186/s13321-020-00464-1
Predicting Target Profiles with Confidence as a Service using Docking Scores
Journal of Cheminformatics. 12, 62 (2020). DOI: 10.1186/s13321-020-00464-1
Norinder U,
Spjuth O,
Svensson F
Using Predicted Bioactivity Profiles to Improve Predictive Modelling
Journal of Chemical Information and Modeling. 60, 6, 2830-2837. (2020). DOI: 10.1021/acs.jcim.0c00250
Using Predicted Bioactivity Profiles to Improve Predictive Modelling
Journal of Chemical Information and Modeling. 60, 6, 2830-2837. (2020). DOI: 10.1021/acs.jcim.0c00250
Capuccini M,
Dahlö M,
Toor S,
and Spjuth O
MaRe: Container-Based Parallel Computing with Data Locality
Gigascience. 9, 5, giaa042. (2020). DOI: 10.1093/gigascience/giaa042
MaRe: Container-Based Parallel Computing with Data Locality
Gigascience. 9, 5, giaa042. (2020). DOI: 10.1093/gigascience/giaa042
Schaduangrat N,
Lampa S,
Simeon S,
Gleeson MP,
Spjuth O,
Nantasenamat C
Towards reproducible computational drug discovery
Journal of Cheminformatics. 12, 9 (2020). DOI: 10.1186/s13321-020-0408-x
Towards reproducible computational drug discovery
Journal of Cheminformatics. 12, 9 (2020). DOI: 10.1186/s13321-020-0408-x
2019
Capuccini M,
Larsson A,
Carone M,
Novella JA,
Sadawi N,
Gao J,
Toor S,
Spjuth O
On-demand virtual research environments using microservices
PeerJ Computer Science. 5, e232 (2019). DOI: 10.7717/peerj-cs.232
On-demand virtual research environments using microservices
PeerJ Computer Science. 5, e232 (2019). DOI: 10.7717/peerj-cs.232
One Thousand Plant Transcriptomes Initiative
One thousand plant transcriptomes and the phylogenomics of green plants
Nature. 574, 679–685. (2019). DOI: 10.1038/s41586-019-1693-2
One thousand plant transcriptomes and the phylogenomics of green plants
Nature. 574, 679–685. (2019). DOI: 10.1038/s41586-019-1693-2
Gauraha,
N.,
Söderdahl,
F. and Spjuth,
O.
Split Knowledge Transfer in Learning Under Privileged Information Framework
Proceedings of Machine Learning Research (PMLR). 105, 43-52. (2019). URL: proceedings.mlr.press/v105/gauraha19a.html
Split Knowledge Transfer in Learning Under Privileged Information Framework
Proceedings of Machine Learning Research (PMLR). 105, 43-52. (2019). URL: proceedings.mlr.press/v105/gauraha19a.html
Spjuth O.,
Brännström R.C.,
Carlsson L. and Gauraha,
N.
Combining Prediction Intervals on Multi-Source Non-Disclosed Regression Datasets
Proceedings of Machine Learning Research (PMLR). 105, 53-65. (2019). URL: proceedings.mlr.press/v105/spjuth19a.html
Combining Prediction Intervals on Multi-Source Non-Disclosed Regression Datasets
Proceedings of Machine Learning Research (PMLR). 105, 53-65. (2019). URL: proceedings.mlr.press/v105/spjuth19a.html
Grüning BA,
Lampa S,
Vaudel M,
Blankenberg D
Software engineering for scientific big data analysis
Gigascience. 8, 5 (2019). DOI: 10.1093/gigascience/giz054
Software engineering for scientific big data analysis
Gigascience. 8, 5 (2019). DOI: 10.1093/gigascience/giz054
Lampa S,
Dahlö M,
Alvarsson J,
Spjuth O
SciPipe-Turning Scientific Workflows into Computer Programs
IEEE Computing in Science & Engineering. 21, 3, 109-113. (2019). DOI: 10.1109/MCSE.2019.2907814
SciPipe-Turning Scientific Workflows into Computer Programs
IEEE Computing in Science & Engineering. 21, 3, 109-113. (2019). DOI: 10.1109/MCSE.2019.2907814
Gupta A,
Harrison PJ,
Wieslander H,
Pielawski N,
Kartasalo K,
Partel G,
Solorzano L,
Suveer A,
Klemm AH,
Spjuth O,
Sintorn I,
Wählby C
Deep Learning in Image Cytometry: A Review
Cytometry. 95, 4 (2019). DOI: 10.1002/cyto.a.23701
Deep Learning in Image Cytometry: A Review
Cytometry. 95, 4 (2019). DOI: 10.1002/cyto.a.23701
Lampa S,
Dahlö M,
Alvarsson J,
Spjuth O
SciPipe: A workflow library for agile development of complex and dynamic bioinformatics pipelines
Gigascience. 8, 5 (2019). DOI: 10.1093/gigascience/giz044
SciPipe: A workflow library for agile development of complex and dynamic bioinformatics pipelines
Gigascience. 8, 5 (2019). DOI: 10.1093/gigascience/giz044
Herman S,
Niemelä V,
Khoonsari PE,
Sundblom J,
Burman J,
Landtblom AM,
Spjuth O,
Nyholm D,
Kultima K
Alterations in the tyrosine and phenylalanine pathways revealed by biochemical profiling in cerebrospinal fluid of Huntington’s disease subjects
Scientific Reports. 9, 4129 (2019). DOI: 10.1038/s41598-019-40186-5
Alterations in the tyrosine and phenylalanine pathways revealed by biochemical profiling in cerebrospinal fluid of Huntington’s disease subjects
Scientific Reports. 9, 4129 (2019). DOI: 10.1038/s41598-019-40186-5
Khoonsari PE,
Moreno P,
Bergmann S,
Burman J,
Capuccini M,
Carone M,
Cascante M,
de Atauri P,
Foguet C,
Gonzalez-Beltran A,
Hankemeier T,
Haug K,
He S,
Herman S,
Johnson D,
Kale N,
Larsson A,
Neumann S,
Peters K,
Pireddu L,
Rocca-Serra P,
Roger P,
Rueedi R,
Ruttkies C,
Sadawi N,
Salek RM,
Sansone SA,
Schober D,
Selivanov V,
Thévenot EA,
van Vliet M,
Zanetti G,
Steinbeck C,
Kultima K,
and Spjuth O
Interoperable and scalable data analysis with microservices: Applications in Metabolomics
Bioinformatics. 35, 19, 3752-3760. (2019). DOI: 10.1093/bioinformatics/btz160
Interoperable and scalable data analysis with microservices: Applications in Metabolomics
Bioinformatics. 35, 19, 3752-3760. (2019). DOI: 10.1093/bioinformatics/btz160
Novella JA,
Khoonsari PE,
Herman S,
Whitenack D,
Capuccini M,
Burman J,
Kultima K,
Spjuth O
Container-based bioinformatics with Pachyderm
Bioinformatics. 35, 5, 839-846. (2019). DOI: 10.1093/bioinformatics/bty699
Container-based bioinformatics with Pachyderm
Bioinformatics. 35, 5, 839-846. (2019). DOI: 10.1093/bioinformatics/bty699
K. Peters,
J. Bradbury,
S. Bergmann,
M. Capuccini,
M. Cascante,
P. de Atauri,
T. M. D. Ebbels,
C. Foguet,
R. Glen,
A. Gonzalez-Beltran,
U. L. Gu ̈nther,
E. Handakas,
T. Hankemeier,
K. Haug,
S. Her- man,
P. Holub,
M. Izzo,
D. Jacob,
D. Johnson,
F. Jourdan,
N. Kale,
I. Karaman,
B. Khalili,
P. E. Khon- sari,
K. Kultima,
S. Lampa,
A. Larsson,
C. Ludwig,
P. Moreno,
S. Neumann,
J. A. Novella,
C. O’Donovan,
J. T. M. Pearce,
A. Peluso,
M. E. Piras,
L. Pireddu,
M. A. C. Reed,
P. Rocca-Serra,
P. Roger,
A. Rosato,
R. Rueedi,
C. Ruttkies,
N. Sadawi,
R. M. Salek,
S.-A. Sansone,
V. Selivanov,
O. Spjuth,
D. Schober,
E. A. Th ́evenot,
M. Tomasoni,
M. van Rijswijk,
M. van Vliet,
M. R. Viant,
R. J. M. Weber,
G. Zanetti,
and C. Steinbeck
PhenoMeNal: Processing and analysis of Metabolomics data in the Cloud
Gigascience. 8, 2, giy149. (2019). DOI: 10.1093/gigascience/giy149
PhenoMeNal: Processing and analysis of Metabolomics data in the Cloud
Gigascience. 8, 2, giy149. (2019). DOI: 10.1093/gigascience/giy149
Herman S,
Åkerfeldt T,
Spjuth O,
Burman J,
Kultima K
Biochemical differences in cerebrospinal fluid between secondary progressive and relapsing-remitting multiple sclerosis
Cells. 8, 2, 84. (2019). DOI: 10.3390/cells8020084
Biochemical differences in cerebrospinal fluid between secondary progressive and relapsing-remitting multiple sclerosis
Cells. 8, 2, 84. (2019). DOI: 10.3390/cells8020084
Kensert A,
Harrison PJ,
Spjuth O
Transfer learning with deep convolutional neural networks for classifying cellular morphological changes
SLAS DISCOVERY: Advancing Life Sciences R&D. 24, 4 (2019). DOI: 10.1177/2472555218818756
Transfer learning with deep convolutional neural networks for classifying cellular morphological changes
SLAS DISCOVERY: Advancing Life Sciences R&D. 24, 4 (2019). DOI: 10.1177/2472555218818756
2018
Lampa S,
Alvarsson J,
Arvidsson Mc Shane S,
Berg A,
Ahlberg E,
Spjuth O
Predicting off-target binding profiles with confidence using Conformal Prediction
Frontiers in Pharmacology. 9, 1256. (2018). DOI: 10.3389/fphar.2018.01256
Predicting off-target binding profiles with confidence using Conformal Prediction
Frontiers in Pharmacology. 9, 1256. (2018). DOI: 10.3389/fphar.2018.01256
Kensert A,
Alvarsson J,
Norinder U,
Spjuth O.
Evaluating parameters for ligand-based modeling with random forest on sparse data sets
Journal of Cheminformatics. 10, 49 (2018). DOI: 10.1186/s13321-018-0304-9
Evaluating parameters for ligand-based modeling with random forest on sparse data sets
Journal of Cheminformatics. 10, 49 (2018). DOI: 10.1186/s13321-018-0304-9
Spjuth O
Novel applications of Machine Learning in cheminformatics
Journal of Cheminformatics. 10, 46 (2018). DOI: 10.1186/s13321-018-0301-z
Novel applications of Machine Learning in cheminformatics
Journal of Cheminformatics. 10, 46 (2018). DOI: 10.1186/s13321-018-0301-z
Spjuth O. and Larsson A.
Deploying PhenoMeNal virtual research environments on the EGI Federated Cloud
Inspired. 31 (2018). URL: www.egi.eu/wp-content/uploads/2018/06/Inspired-Issue-31.pdf
Deploying PhenoMeNal virtual research environments on the EGI Federated Cloud
Inspired. 31 (2018). URL: www.egi.eu/wp-content/uploads/2018/06/Inspired-Issue-31.pdf
Herman S,
Khoonsari PE,
Tolf A,
Steinmetz J,
Zetterberg H,
Åkerfeldt T,
Jakobsson P-J,
Larsson A,
Spjuth O,
Burman J,
Kultima K
Integration of Magnetic Resonance Imaging and Protein and Metabolite CSF Measurements to Enable Early Diagnosis of Secondary Progressive Multiple Sclerosis
Theranostics. 8, 16, 4477-4490. (2018). DOI: 10.7150/thno.26249
Integration of Magnetic Resonance Imaging and Protein and Metabolite CSF Measurements to Enable Early Diagnosis of Secondary Progressive Multiple Sclerosis
Theranostics. 8, 16, 4477-4490. (2018). DOI: 10.7150/thno.26249
Gauraha N,
Carlsson L,
Spjuth O.
Conformal Prediction in Learning Under Privileged Information Paradigm with Applications in Drug Discovery.
Proceedings of Machine Learning Research. 91, 147-156 (2018). URL: proceedings.mlr.press/v91/gauraha18a.html
Conformal Prediction in Learning Under Privileged Information Paradigm with Applications in Drug Discovery.
Proceedings of Machine Learning Research. 91, 147-156 (2018). URL: proceedings.mlr.press/v91/gauraha18a.html
Svensson F,
Aniceto N,
Norinder U,
Cortes I,
Spjuth O,
Carlsson L,
Bender A.
Conformal Regression for QSAR Modelling - Quantifying Prediction Uncertainty.
Journal of Chemical Information and Modeling. 58, 5, 1132–1140. (2018). DOI: 10.1021/acs.jcim.8b00054
Conformal Regression for QSAR Modelling - Quantifying Prediction Uncertainty.
Journal of Chemical Information and Modeling. 58, 5, 1132–1140. (2018). DOI: 10.1021/acs.jcim.8b00054
Lapins M,
Arvidsson S,
Lampa S,
Berg A,
Schaal W,
Alvarsson J,
Spjuth O.
A confidence predictor for logD using conformal regression and a support-vector machine.
Journal of Cheminformatics. 10, 18 (2018). DOI: 10.1186/s13321-018-0271-1
A confidence predictor for logD using conformal regression and a support-vector machine.
Journal of Cheminformatics. 10, 18 (2018). DOI: 10.1186/s13321-018-0271-1
Ahmed L,
Georgiev V,
Capuccini M,
Toor S,
Schaal W,
Laure E,
Spjuth O.
Efficient Iterative Virtual Screening with Apache Spark and Conformal Prediction
Journal of Cheminformatics. 10, 8 (2018). DOI: 10.1186/s13321-018-0265-z
Efficient Iterative Virtual Screening with Apache Spark and Conformal Prediction
Journal of Cheminformatics. 10, 8 (2018). DOI: 10.1186/s13321-018-0265-z
Dahlö M,
Scofield DG,
Schaal W,
Spjuth O.
Tracking the NGS revolution: Managing life science research on shared high-performance computing clusters.
GigaScience. 7, 5 (2018). DOI: 10.1093/gigascience/giy028
Tracking the NGS revolution: Managing life science research on shared high-performance computing clusters.
GigaScience. 7, 5 (2018). DOI: 10.1093/gigascience/giy028
2017
van Rijswijk M,
Beirnaert C,
Caron C,
Cascante M,
Dominguez V,
Dunn W,
Ebbels T,
Giacomoni F,
Gonzalez-Beltran A,
Hankemeier T,
Haug K,
Izquierdo-Garcia J,
Jimenez R,
Jourdan F,
Kale N,
Klapa M,
Kohlbacher O,
Koort K,
Kultima K,
Le Corguill G,
Moschonas N,
Neumann S,
O'Donovan C,
Reczko M,
Rocca-Serra P,
Rosato A,
Salek R,
Sansone S,
Satagopam V,
Schober D,
Shimmo R,
Spicer R,
Spjuth O,
Thevenot E,
Viant M,
Weber R,
Willighagen E,
Zanetti G,
and Steinbeck C
The future of metabolomics in ELIXIR
F1000Research. 6(ELIXIR), 1649 (2017). DOI: 10.12688/f1000research.12342.2
The future of metabolomics in ELIXIR
F1000Research. 6(ELIXIR), 1649 (2017). DOI: 10.12688/f1000research.12342.2
Lampa S,
Willighagen E,
Kohonen P,
King A,
Vrandečić D,
Grafström R,
Spjuth O
RDFIO: extending Semantic MediaWiki for interoperable biomedical data management
Journal of Biomedical Semantics. 8, 35 (2017). DOI: 10.1186/s13326-017-0136-y
RDFIO: extending Semantic MediaWiki for interoperable biomedical data management
Journal of Biomedical Semantics. 8, 35 (2017). DOI: 10.1186/s13326-017-0136-y
Spjuth O,
Karlsson A,
Clements M,
Humphreys K,
Ivansson E,
Dowling J,
Eklund M,
Jauhiainen A,
Czene K,
Grönberg H,
Sparén P,
Wiklund F,
Cheddad A,
Pálsdóttir þ,
Rantalainen M,
Abrahamsson L,
Laure E,
Litton J-E,
Palmgren J
E-Science technologies in a workflow for personalized medicine using cancer screening as a case study
Journal of the American Medical Informatics Association. 24, 5, 950-957. (2017). DOI: 10.1093/jamia/ocx038
E-Science technologies in a workflow for personalized medicine using cancer screening as a case study
Journal of the American Medical Informatics Association. 24, 5, 950-957. (2017). DOI: 10.1093/jamia/ocx038
Toor S,
Lindberg M,
Fallman I,
Vallin A,
Mohill O,
Freyhult P,
Nilsson L,
Agback M,
Viklund L,
Zazzi H,
Spjuth O,
Capuccini M,
Moller J,
Murtagh D,
Hellander A
SNIC Science Cloud (SSC): A National-scale Cloud Infrastructure for Swedish Academia
e-Science (e-Science), 2017 IEEE 13th International Conference on. 10, 219-227. (2017). DOI: 10.1109/eScience.2017.35
SNIC Science Cloud (SSC): A National-scale Cloud Infrastructure for Swedish Academia
e-Science (e-Science), 2017 IEEE 13th International Conference on. 10, 219-227. (2017). DOI: 10.1109/eScience.2017.35
Sütterlin S,
Dahlö M,
Tellgren-Roth C,
Schaal W,
Melhus Å
High frequency of silver resistance genes in invasive isolates of Enterobacter and Klebsiella species
J Hosp Infect. 96, 3, 256-261. (2017). DOI: 10.1016/j.jhin.2017.04.017
High frequency of silver resistance genes in invasive isolates of Enterobacter and Klebsiella species
J Hosp Infect. 96, 3, 256-261. (2017). DOI: 10.1016/j.jhin.2017.04.017
Arvidsson S,
Carlsson L,
Toccaceli P,
and Spjuth O.
Prediction of Metabolic Transformations using Cross Venn-ABERS Predictors
Proceedings of Machine Learning Research. 60, 1-14. (2017). URL: proceedings.mlr.press/v60/arvidsson17a.html
Prediction of Metabolic Transformations using Cross Venn-ABERS Predictors
Proceedings of Machine Learning Research. 60, 1-14. (2017). URL: proceedings.mlr.press/v60/arvidsson17a.html
Willighagen E,
Mayfield J,
Alvarsson J,
Berg A,
Carlsson L,
Jeliazkova N,
Kuhn S,
Pluskal T,
Rojas-Chertó M,
Spjuth O,
Torrance G,
Evelo C,
Guha R,
Steinbeck C
The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching
Journal of Cheminformatics. 9, 33 (2017). DOI: 10.1186/s13321-017-0220-4
The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching
Journal of Cheminformatics. 9, 33 (2017). DOI: 10.1186/s13321-017-0220-4
Herman S,
Khoonsari PE,
Aftab O,
Krishnan S,
Strömbom E,
Larsson R,
Hammerling U,
Spjuth O,
Kultima M,
Gustafsson M
Mass spectrometry based metabolomics for in vitro systems pharmacology: pitfalls, challenges, and computational solutions
Metabolomics. 13, 79 (2017). DOI: 10.1007/s11306-017-1213-z
Mass spectrometry based metabolomics for in vitro systems pharmacology: pitfalls, challenges, and computational solutions
Metabolomics. 13, 79 (2017). DOI: 10.1007/s11306-017-1213-z
Shoombuatong W,
Prathipati P,
Prachayasittikul V,
Schaduangrat N,
Malik AA,
Pratiwi R,
Wanwimolruk S,
Wikberg JES,
Gleeson MP,
Spjuth O,
Nantasenamat C
Towards Predicting the Cytochrome P450 Modulation: From QSAR to proteochemometric modeling
Current Drug Metabolism. 18, 6, 540-555. (2017). DOI: 10.2174/1389200218666170320121932
Towards Predicting the Cytochrome P450 Modulation: From QSAR to proteochemometric modeling
Current Drug Metabolism. 18, 6, 540-555. (2017). DOI: 10.2174/1389200218666170320121932
Capuccini M,
Ahmed L,
Schaal W,
Laure E,
Spjuth O
Large-scale virtual screening on public cloud resources with Apache Spark
Journal of Cheminformatics. 9, 15. (2017). DOI: 10.1186/s13321-017-0204-4
Large-scale virtual screening on public cloud resources with Apache Spark
Journal of Cheminformatics. 9, 15. (2017). DOI: 10.1186/s13321-017-0204-4
Alogheli H,
Olanders G,
Schaal W,
Brandt P,
Karlén A
Docking of Macrocycles: Comparing Rigid and Flexible Docking in Glide
J Chem Inf Model. 57, 2, 190-202. (2017). DOI: 10.1021/acs.jcim.6b00443
Docking of Macrocycles: Comparing Rigid and Flexible Docking in Glide
J Chem Inf Model. 57, 2, 190-202. (2017). DOI: 10.1021/acs.jcim.6b00443
2016
Lampa S,
Alvarsson J,
Spjuth O
Towards agile large-scale predictive modelling in drug discovery with flow-based programming design principles
Journal of Cheminformatics. 8, 67. (2016). DOI: 10.1186/s13321-016-0179-6
Towards agile large-scale predictive modelling in drug discovery with flow-based programming design principles
Journal of Cheminformatics. 8, 67. (2016). DOI: 10.1186/s13321-016-0179-6
Spjuth O,
Rydberg P,
Willighagen EL,
Evelo CT,
Jeliazkova N
XMetDB: an open access database for xenobiotic metabolism
Journal of Cheminformatics. 8, 47. (2016). DOI: 10.1186/s13321-016-0161-3
XMetDB: an open access database for xenobiotic metabolism
Journal of Cheminformatics. 8, 47. (2016). DOI: 10.1186/s13321-016-0161-3
Alvarsson J,
Lampa S,
Schaal W,
Andersson C,
Wikberg JES,
Spjuth O
Large-scale ligand-based predictive modelling using support vector machines
Journal of Cheminformatics. 8, 39. (2016). DOI: 10.1186/s13321-016-0151-5
Large-scale ligand-based predictive modelling using support vector machines
Journal of Cheminformatics. 8, 39. (2016). DOI: 10.1186/s13321-016-0151-5
Spjuth O,
Bongcam-Rudloff E,
Dahlberg J,
Dahlö M,
Kallio A,
Pireddu L,
Vezzi F,
Korpelainen E
Recommendations on e-infrastructures for next-generation sequencing
GigaScience. 5, 1 (2016). DOI: 10.1186/s13742-016-0132-7
Recommendations on e-infrastructures for next-generation sequencing
GigaScience. 5, 1 (2016). DOI: 10.1186/s13742-016-0132-7
Simeon S,
Spjuth O,
Lapins M,
Nabu S,
Anuwongcharoen N,
Prachayasittikul V,
Wikberg JES,
Nantasenamat C
Origin of aromatase inhibitory activity via proteochemometric modeling
PeerJ. 4, e1979. (2016). DOI: 10.7717/peerj.1979
Origin of aromatase inhibitory activity via proteochemometric modeling
PeerJ. 4, e1979. (2016). DOI: 10.7717/peerj.1979
Spjuth O,
Krestyaninova M,
Hastings J,
Shen H-Y,
Heikkinen J,
Waldenberger M,
Langhammer A,
Ladenvall C,
Esko T,
Persson M-Å,
Heggland J,
Dietrich J,
Ose S,
Gieger C,
Ried JS,
Peters A,
Fortier I,
de Geus EJC,
Klovins J,
Zaharenko L,
Willemsen G,
Hottenga J-J,
Litton J-E,
Karvanen J,
Boomsma DI,
Groop L,
Rung J,
Palmgren J,
Pedersen NL,
McCarthy MI,
van Duijn CM,
Hveem K,
Metspalu A,
Ripatti S,
Prokopenko I,
Harris JR
Harmonising and linking biomedical and clinical data across disparate data archives to enable integrative cross-biobank research
Eur J Hum Genet. 24, 4, 521--528. (2016). DOI: 10.1038/ejhg.2015.165
Harmonising and linking biomedical and clinical data across disparate data archives to enable integrative cross-biobank research
Eur J Hum Genet. 24, 4, 521--528. (2016). DOI: 10.1038/ejhg.2015.165
2015
Capuccini M,
Carlsson L,
Norinder U,
Spjuth O
Conformal Prediction in Spark: Large-Scale Machine Learning with Confidence
Proceedings - 2015 2nd IEEE/ACM International Symposium on Big Data Computing, BDC 2015. , 61-67. (2015). DOI: 10.1109/BDC.2015.35
Conformal Prediction in Spark: Large-Scale Machine Learning with Confidence
Proceedings - 2015 2nd IEEE/ACM International Symposium on Big Data Computing, BDC 2015. , 61-67. (2015). DOI: 10.1109/BDC.2015.35
Grafström RC,
Nymark P,
Hongisto V,
Spjuth O,
Ceder R,
Willighagen E,
Hardy B,
Kaski S,
Kohonen P
Toward the Replacement of Animal Experiments through the Bioinformatics-driven Analysis of 'Omics' Data from Human Cell Cultures
Altern. Lab. Anim.. 43, 5, 325--332. (2015).
Toward the Replacement of Animal Experiments through the Bioinformatics-driven Analysis of 'Omics' Data from Human Cell Cultures
Altern. Lab. Anim.. 43, 5, 325--332. (2015).
Dahlö M,
Haziza F,
Kallio A,
Korpelainen E,
Bongcam-Rudloff E,
Spjuth O
BioImg.org: A catalog of virtual machine images for the life sciences
Bioinformatics and Biology Insights. 9, 125-128. (2015). DOI: 10.4137/BBI.S28636
BioImg.org: A catalog of virtual machine images for the life sciences
Bioinformatics and Biology Insights. 9, 125-128. (2015). DOI: 10.4137/BBI.S28636
Spjuth O,
Bongcam-Rudloff E,
Hernández GC,
Forer L,
Giovacchini M,
Guimera RV,
Kallio A,
Korpelainen E,
Kańduła MM,
Krachunov M,
Kreil DP,
Kulev O,
Łabaj PP,
Lampa S,
Pireddu L,
Schönherr S,
Siretskiy A,
Vassilev D
Experiences with workflows for automating data-intensive bioinformatics
Biology Direct. 10, 1 (2015). DOI: 10.1186/s13062-015-0071-8
Experiences with workflows for automating data-intensive bioinformatics
Biology Direct. 10, 1 (2015). DOI: 10.1186/s13062-015-0071-8
Blom K,
Nygren P,
Alvarsson J,
Larsson R,
Andersson CR
Ex Vivo Assessment of Drug Activity in Patient Tumor Cells as a Basis for Tailored Cancer Therapy
Journal of Laboratory Automation. 21, 1, 178-187. (2015). DOI: 10.1177/2211068215598117
Ex Vivo Assessment of Drug Activity in Patient Tumor Cells as a Basis for Tailored Cancer Therapy
Journal of Laboratory Automation. 21, 1, 178-187. (2015). DOI: 10.1177/2211068215598117
Siretskiy A,
Sundqvist T,
Voznesenskiy M,
Spjuth O
A quantitative assessment of the Hadoop framework for analyzing massively parallel DNA sequencing data
GigaScience. 4, 1 (2015). DOI: 10.1186/s13742-015-0058-5
A quantitative assessment of the Hadoop framework for analyzing massively parallel DNA sequencing data
GigaScience. 4, 1 (2015). DOI: 10.1186/s13742-015-0058-5
Gholami A,
Laure E,
Somogyi P,
Spjuth O,
Niazi S,
Dowling J
Privacy-Preservation for Publishing Sample Availability Data with Personal Identifiers
JOMB. 4, 2, 117--125. (2015). DOI: 10.12720/jomb.4.2.117-125
Privacy-Preservation for Publishing Sample Availability Data with Personal Identifiers
JOMB. 4, 2, 117--125. (2015). DOI: 10.12720/jomb.4.2.117-125
Ahlberg E,
Spjuth O,
Hasselgren C,
Carlsson L
Interpretation of conformal prediction classification models
Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 9047, 323-334. (2015). DOI: 10.1007/978-3-319-17091-6_27
Interpretation of conformal prediction classification models
Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 9047, 323-334. (2015). DOI: 10.1007/978-3-319-17091-6_27
Lindh M,
Svensson F,
Schaal W,
Zhang J,
Sköld C,
Brandt P,
Karlén A
Toward a benchmarking data set able to evaluate ligand- and structure-based virtual screening using public HTS data
J Chem Inf Model. 55, 2, 343-353. (2015). DOI: 10.1021/ci5005465
Toward a benchmarking data set able to evaluate ligand- and structure-based virtual screening using public HTS data
J Chem Inf Model. 55, 2, 343-353. (2015). DOI: 10.1021/ci5005465
Moghadam BT,
Alvarsson J,
Holm M,
Eklund M,
Carlsson L,
Spjuth O
Scaling predictive modeling in drug development with cloud computing
Journal of Chemical Information and Modeling. 55, 1, 19-25. (2015). DOI: 10.1021/ci500580y
Scaling predictive modeling in drug development with cloud computing
Journal of Chemical Information and Modeling. 55, 1, 19-25. (2015). DOI: 10.1021/ci500580y
2014
Siretskiy A,
Spjuth O
HTSeq-Hadoop: Extending HTSeq for massively parallel sequencing data analysis using Hadoop
Proceedings - 2014 IEEE 10th International Conference on eScience, eScience 2014. 1, 317-323. (2014). DOI: 10.1109/eScience.2014.27
HTSeq-Hadoop: Extending HTSeq for massively parallel sequencing data analysis using Hadoop
Proceedings - 2014 IEEE 10th International Conference on eScience, eScience 2014. 1, 317-323. (2014). DOI: 10.1109/eScience.2014.27
Alvarsson J,
Eklund M,
Andersson C,
Carlsson L,
Spjuth O,
Wikberg JES
Benchmarking Study of Parameter Variation When Using Signature Fingerprints Together with Support Vector Machines
Journal of Chemical Information and Modeling. 54, 11, 3211--3217. (2014). DOI: 10.1021/ci500344v
Benchmarking Study of Parameter Variation When Using Signature Fingerprints Together with Support Vector Machines
Journal of Chemical Information and Modeling. 54, 11, 3211--3217. (2014). DOI: 10.1021/ci500344v
Alvarsson J,
Eklund M,
Engkvist O,
Spjuth O,
Carlsson L,
Wikberg JES,
Noeske T
Ligand-based target prediction with signature fingerprints
Journal of Chemical Information and Modeling. 54, 10, 2647–2653. (2014). DOI: 10.1021/ci500361u
Ligand-based target prediction with signature fingerprints
Journal of Chemical Information and Modeling. 54, 10, 2647–2653. (2014). DOI: 10.1021/ci500361u
Spjuth O,
Heikkinen J,
Litton J-E,
Palmgren J,
Krestyaninova M
Data integration between Swedish national clinical health registries and biobanks using an availability system
Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 8574 LNBI, 32-40. (2014). DOI: 10.1007/978-3-319-08590-6_3
Data integration between Swedish national clinical health registries and biobanks using an availability system
Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 8574 LNBI, 32-40. (2014). DOI: 10.1007/978-3-319-08590-6_3
Spjuth O
NGS data management and analysis for hundreds of projects: Experiences from Sweden
EMBnet.journal. 20, A, 761. (2014). DOI: 10.14806/ej.20.a.761
NGS data management and analysis for hundreds of projects: Experiences from Sweden
EMBnet.journal. 20, A, 761. (2014). DOI: 10.14806/ej.20.a.761
Kohonen P,
Ceder R,
Smit I,
Hongisto V,
Myatt G,
Hardy B,
Spjuth O,
Grafström R
Cancer biology, toxicology and alternative methods development go hand-in-hand.
Basic & Clinical Pharmacology & Toxicology. 15, 1, 50-58. (2014). DOI: 10.1111/bcpt.12257
Cancer biology, toxicology and alternative methods development go hand-in-hand.
Basic & Clinical Pharmacology & Toxicology. 15, 1, 50-58. (2014). DOI: 10.1111/bcpt.12257
2013
Ahmed L,
Edlund A,
Laure E,
Spjuth O
Using iterative MapReduce for parallel virtual screening
Proceedings of the International Conference on Cloud Computing Technology and Science, CloudCom. 2, 27-32. (2013). DOI: 10.1109/CloudCom.2013.99
Using iterative MapReduce for parallel virtual screening
Proceedings of the International Conference on Cloud Computing Technology and Science, CloudCom. 2, 27-32. (2013). DOI: 10.1109/CloudCom.2013.99
Spjuth O,
Carlsson L,
Alvarsson J,
Georgiev V,
Willighagen E,
Eklund M
Open Source Drug Discovery with Bioclipse
Current Topics in Medicinal Chemistry. 12, 18 (2013). DOI: 10.2174/1568026611212180005
Open Source Drug Discovery with Bioclipse
Current Topics in Medicinal Chemistry. 12, 18 (2013). DOI: 10.2174/1568026611212180005
Schaal W,
Hammerling U,
Gustafsson MG,
Spjuth O
Automated QuantMap for rapid quantitative molecular network topology analysis
Bioinformatics. 29, 18, 2369-2370. (2013). DOI: 10.1093/bioinformatics/btt390
Automated QuantMap for rapid quantitative molecular network topology analysis
Bioinformatics. 29, 18, 2369-2370. (2013). DOI: 10.1093/bioinformatics/btt390
Lampa S,
Dahlö M,
Olason PI,
Hagberg J,
Spjuth O
Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data
GigaScience. 2, 1 (2013). DOI: 10.1186/2047-217x-2-9
Lessons learned from implementing a national infrastructure in Sweden for storage and analysis of next-generation sequencing data
GigaScience. 2, 1 (2013). DOI: 10.1186/2047-217x-2-9
Lapins M,
Worachartcheewan A,
Spjuth O,
Georgiev V,
Prachayasittikul V,
Nantasenamat C,
Wikberg JES
A Unified Proteochemometric Model for Prediction of Inhibition of Cytochrome P450 Isoforms
PLoS One. 8, 6, e66566. (2013). DOI: 10.1371/journal.pone.0066566
A Unified Proteochemometric Model for Prediction of Inhibition of Cytochrome P450 Isoforms
PLoS One. 8, 6, e66566. (2013). DOI: 10.1371/journal.pone.0066566
Rostkowski M,
Spjuth O,
Rydberg P
WhichCyp: prediction of cytochromes P450 inhibition.
Bioinformatics. 29, 16, 2051-2052. (2013). DOI: 10.1093/bioinformatics/btt325
WhichCyp: prediction of cytochromes P450 inhibition.
Bioinformatics. 29, 16, 2051-2052. (2013). DOI: 10.1093/bioinformatics/btt325
Willighagen EL,
Waagmeester A,
Spjuth O,
Ansell P,
Williams AJ,
Tkachenko V,
Hastings J,
Chen B,
Wild DJ
The ChEMBL database as linked open data
Journal of Cheminformatics. 5, 23 (2013). DOI: 10.1186/1758-2946-5-23
The ChEMBL database as linked open data
Journal of Cheminformatics. 5, 23 (2013). DOI: 10.1186/1758-2946-5-23
Claesson A,
Spjuth O
On mechanisms of reactive metabolite formation from drugs.
Mini Rev Med Chem. 13, 5, 720-9. (2013).
On mechanisms of reactive metabolite formation from drugs.
Mini Rev Med Chem. 13, 5, 720-9. (2013).
Spjuth O,
Berg A,
Adams S,
Willighagen EL
Applications of the InChI in cheminformatics with the CDK and Bioclipse
Journal of Cheminformatics. 5, 3 (2013). DOI: 10.1186/1758-2946-5-14
Applications of the InChI in cheminformatics with the CDK and Bioclipse
Journal of Cheminformatics. 5, 3 (2013). DOI: 10.1186/1758-2946-5-14
2012
Spjuth O,
Georgiev V,
Carlsson L,
Alvarsson J,
Berg A,
Willighagen E,
Wikberg JES,
Eklund M
Bioclipse-R: integrating management and visualization of life science data with statistical analysis.
Bioinformatics. 29, 2, 286-289. (2012). DOI: 10.1093/bioinformatics/bts681
Bioclipse-R: integrating management and visualization of life science data with statistical analysis.
Bioinformatics. 29, 2, 286-289. (2012). DOI: 10.1093/bioinformatics/bts681
Spjuth O,
Carlsson L,
Alvarsson J,
Georgiev V,
Willighagen E,
Eklund M
Open source drug discovery with Bioclipse
Current Topics in Medicinal Chemistry. 12, 18, 1980-1986. (2012). DOI: 10.2174/156802612804910287
Open source drug discovery with Bioclipse
Current Topics in Medicinal Chemistry. 12, 18, 1980-1986. (2012). DOI: 10.2174/156802612804910287
Williams AJ,
Ekins S,
Spjuth O,
Willighagen EL
Accessing, using, and creating chemical property databases for computational toxicology modeling
Computational Toxicology, Methods in Molecular Biology. 929, 221-241. (2012). DOI: 10.1007/978-1-62703-050-2_10
Accessing, using, and creating chemical property databases for computational toxicology modeling
Computational Toxicology, Methods in Molecular Biology. 929, 221-241. (2012). DOI: 10.1007/978-1-62703-050-2_10
Willighagen E,
Affentranger R,
Grafström R,
Hardy B,
Jeliazkova N,
Spjuth O
Interactive predictive toxicology with Bioclipse and OpenTox
Open Source Software in Life Science Research. , 35--61. (2012). DOI: 10.1533/9781908818249.35
Interactive predictive toxicology with Bioclipse and OpenTox
Open Source Software in Life Science Research. , 35--61. (2012). DOI: 10.1533/9781908818249.35
Hardy B,
Apic G,
Carthew P,
Clark D,
Cook D,
Escher S,
Dix I,
Hastings J,
Heard DJ,
Jeliazkova N,
Judson P,
Matis-Mitchell S,
Mitic D,
Myatt G,
Shah I,
Spjuth O,
Tcheremenskaia O,
Toldo L,
Watson D,
White A,
Yang C
Food for thought ... A toxicology ontology roadmap
ALTEX. 29, 2, 127-138. (2012). DOI: 10.14573/altex.2012.2.129
Food for thought ... A toxicology ontology roadmap
ALTEX. 29, 2, 127-138. (2012). DOI: 10.14573/altex.2012.2.129
Carlsson L,
Spjuth O,
Eklund M,
Boyer S
Model building in Bioclipse Decision Support applied to open datasets
Toxicology Letters. 211, S62. (2012). DOI: 10.1016/j.toxlet.2012.03.243
Model building in Bioclipse Decision Support applied to open datasets
Toxicology Letters. 211, S62. (2012). DOI: 10.1016/j.toxlet.2012.03.243
Spjuth O,
Willighagen E,
Hammerling U,
Dencker L,
Grafström R
A novel infrastructure for chemical safety predictions with focus on human health
Toxicology Letters. 211, S59. (2012). DOI: 10.1016/j.toxlet.2012.03.234
A novel infrastructure for chemical safety predictions with focus on human health
Toxicology Letters. 211, S59. (2012). DOI: 10.1016/j.toxlet.2012.03.234
Hardy B,
Apic G,
Carthew P,
Clark D,
Cook D,
Dix I,
Escher S,
Hastings J,
Heard DJ,
Jeliazkova N,
Judson P,
Matis-Mitchell S,
Mitic D,
Myatt G,
Shah I,
Spjuth O,
Tcheremenskaia O,
Toldo L,
Watson D,
White A,
Yang C
Toxicology Ontology Perspectives
ALTEX. 29, 2, 139-156. (2012). DOI: 10.14573/altex.2012.2.139
Toxicology Ontology Perspectives
ALTEX. 29, 2, 139-156. (2012). DOI: 10.14573/altex.2012.2.139
Lampa S,
Hagberg J,
Spjuth O
UPPNEX - A solution for Next Generation Sequencing data management and analysis
EMBnet journal. 17, B, 44. (2012). DOI: 10.14806/ej.17.b.274
UPPNEX - A solution for Next Generation Sequencing data management and analysis
EMBnet journal. 17, B, 44. (2012). DOI: 10.14806/ej.17.b.274
Krestyaninova M,
Spjuth O,
Hastings J,
Dietrich J,
Rebholz-Schuhmann D
Biobank Metaportal to Enhance Collaborative Research: sail.simbioms.org
Journal of Systemics, Cybernetics and Informatics. 10 (2012). URL: www.iiisci.org/journal/sci/Abstract.asp?var=&id=HCT875XS
Biobank Metaportal to Enhance Collaborative Research: sail.simbioms.org
Journal of Systemics, Cybernetics and Informatics. 10 (2012). URL: www.iiisci.org/journal/sci/Abstract.asp?var=&id=HCT875XS
2011
Wikberg JES,
Spjuth O,
Eklund M,
Lapins M
Chemoinformatics Taking Biology into Account: Proteochemometrics
Computational Approaches in Cheminformatics and Bioinformatics. , 57-92. (2011). DOI: 10.1002/9781118131411.ch3
Chemoinformatics Taking Biology into Account: Proteochemometrics
Computational Approaches in Cheminformatics and Bioinformatics. , 57-92. (2011). DOI: 10.1002/9781118131411.ch3
Willighagen EL,
Jeliazkova N,
Hardy B,
Grafström RC,
Spjuth O
Computational toxicology using the OpenTox application programming interface and Bioclipse
BMC Research Notes. 4 (2011). DOI: 10.1186/1756-0500-4-487
Computational toxicology using the OpenTox application programming interface and Bioclipse
BMC Research Notes. 4 (2011). DOI: 10.1186/1756-0500-4-487
O'Boyle NM,
Guha R,
Willighagen EL,
Adams SE,
Alvarsson J,
Bradley,
J-C,
Filippov IV,
Hanson RM,
Hanwell MD,
Hutchison GR,
James CA,
Jeliazkova N,
Lang ASID,
Langner KM,
Lonie DC,
Lowe DM,
Pansanel,
Jérôme,
Pavlov D,
Spjuth O,
Steinbeck C,
Tenderholt AL,
Theisen KJ,
Murray-Rust P
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk five years on.
Journal of Cheminformatics. 3, 37 (2011). DOI: 10.1186/1758-2946-3-37
Open Data, Open Source and Open Standards in chemistry: The Blue Obelisk five years on.
Journal of Cheminformatics. 3, 37 (2011). DOI: 10.1186/1758-2946-3-37
Spjuth O,
Eklund M,
Ahlberg Helgee E,
Boyer S,
Carlsson L
Integrated decision support for assessing chemical liabilities
Journal of Chemical Information and Modeling. 51, 8, 1840-1847. (2011). DOI: 10.1021/ci200242c
Integrated decision support for assessing chemical liabilities
Journal of Chemical Information and Modeling. 51, 8, 1840-1847. (2011). DOI: 10.1021/ci200242c
Alvarsson J,
Andersson C,
Spjuth O,
Larsson R,
Wikberg JES
Brunn: An open source laboratory information system for microplates with a graphical plate layout design process
BMC Bioinformatics. 12 (2011). DOI: 10.1186/1471-2105-12-179
Brunn: An open source laboratory information system for microplates with a graphical plate layout design process
BMC Bioinformatics. 12 (2011). DOI: 10.1186/1471-2105-12-179
Guha R,
Spjuth O,
Willighagen E
Collaborative Cheminformatics Applications
Collaborative Computational Technologies for Biomedical Research. , 399-422. (2011). DOI: 10.1002/9781118026038.ch24
Collaborative Cheminformatics Applications
Collaborative Computational Technologies for Biomedical Research. , 399-422. (2011). DOI: 10.1002/9781118026038.ch24
Spjuth O,
Eklund M,
Lapins M,
Junaid M,
Wikberg JES
Services for prediction of drug susceptibility for HIV proteases and reverse transcriptases at the HIV drug research centre
Bioinformatics. 27, 12, 1719-1720. (2011). DOI: 10.1093/bioinformatics/btr192
Services for prediction of drug susceptibility for HIV proteases and reverse transcriptases at the HIV drug research centre
Bioinformatics. 27, 12, 1719-1720. (2011). DOI: 10.1093/bioinformatics/btr192
Willighagen EL,
Alvarsson J,
Andersson A,
Eklund M,
Lampa S,
Lapins M,
Spjuth O,
Wikberg JES
Linking the Resource Description Framework to cheminformatics and proteochemometrics
Journal of Biomedical Semantics. 2, 1 (2011). DOI: 10.1186/2041-1480-2-S1-S6
Linking the Resource Description Framework to cheminformatics and proteochemometrics
Journal of Biomedical Semantics. 2, 1 (2011). DOI: 10.1186/2041-1480-2-S1-S6
2010
Spjuth O
Proteochemometric modeling of the susceptibility of mutated variants of the HIV-1 virus to reverse transcriptase inhibitors
PLoS One. 5, 12 (2010). DOI: 10.1371/journal.pone.0014353
Proteochemometric modeling of the susceptibility of mutated variants of the HIV-1 virus to reverse transcriptase inhibitors
PLoS One. 5, 12 (2010). DOI: 10.1371/journal.pone.0014353
Carlsson L,
Spjuth O,
Adams S,
Glen RC,
Boyer S
Use of historic metabolic biotransformation data as a means of anticipating metabolic sites using MetaPrint2D and Bioclipse
BMC Bioinformatics. 11 (2010). DOI: 10.1186/1471-2105-11-362
Use of historic metabolic biotransformation data as a means of anticipating metabolic sites using MetaPrint2D and Bioclipse
BMC Bioinformatics. 11 (2010). DOI: 10.1186/1471-2105-11-362
Spjuth O,
Willighagen EL,
Guha R,
Eklund M,
Wikberg JES
Towards interoperable and reproducible QSAR analyses: Exchange of datasets
Journal of Cheminformatics. 2, 1 (2010). DOI: 10.1186/1758-2946-2-5
Towards interoperable and reproducible QSAR analyses: Exchange of datasets
Journal of Cheminformatics. 2, 1 (2010). DOI: 10.1186/1758-2946-2-5
Eklund M,
Spjuth O,
Wikberg JES
An eScience-Bayes strategy for analyzing omics data
BMC Bioinformatics. 11 (2010). DOI: 10.1186/1471-2105-11-282
An eScience-Bayes strategy for analyzing omics data
BMC Bioinformatics. 11 (2010). DOI: 10.1186/1471-2105-11-282
2009
Spjuth O,
Alvarsson J,
Berg A,
Eklund M,
Kuhn S,
Mäsak C,
Torrance G,
Wagener J,
Willighagen EL,
Steinbeck C,
Wikberg JES
Bioclipse 2: A scriptable integration platform for the life sciences
BMC Bioinformatics. 10 (2009). DOI: 10.1186/1471-2105-10-397
Bioclipse 2: A scriptable integration platform for the life sciences
BMC Bioinformatics. 10 (2009). DOI: 10.1186/1471-2105-10-397
Wagener J,
Spjuth O,
Willighagen EL,
Wikberg JES
XMPP for cloud computing in bioinformatics supporting discovery and invocation of asynchronous web services
BMC Bioinformatics. 10, 279. (2009). DOI: 10.1186/1471-2105-10-279
XMPP for cloud computing in bioinformatics supporting discovery and invocation of asynchronous web services
BMC Bioinformatics. 10, 279. (2009). DOI: 10.1186/1471-2105-10-279
2008
Eklund M,
Spjuth O,
Wikberg JES
The C1C2: A framework for simultaneous model selection and assessment
BMC Bioinformatics. 9 (2008). DOI: 10.1186/1471-2105-9-360
The C1C2: A framework for simultaneous model selection and assessment
BMC Bioinformatics. 9 (2008). DOI: 10.1186/1471-2105-9-360
Lapins M,
Eklund M,
Spjuth O,
Prusis P,
Wikberg JES
Proteochemometric modeling of HIV protease susceptibility
BMC Bioinformatics. 9 (2008). DOI: 10.1186/1471-2105-9-181
Proteochemometric modeling of HIV protease susceptibility
BMC Bioinformatics. 9 (2008). DOI: 10.1186/1471-2105-9-181
2007
Spjuth O,
Helmus T,
Willighagen EL,
Kuhn S,
Eklund M,
Wagener J,
Murray-Rust P,
Steinbeck C,
Wikberg JES
Bioclipse: An open source workbench for chemo- and bioinformatics
BMC Bioinformatics. 8 (2007). DOI: 10.1186/1471-2105-8-59
Bioclipse: An open source workbench for chemo- and bioinformatics
BMC Bioinformatics. 8 (2007). DOI: 10.1186/1471-2105-8-59
2006
Ameur A,
Yankovski V,
Enroth S,
Spjuth O,
Komorowski J.
The LCB Data Warehouse
Bioinformatics. 22, 8, 1024-6. (2006). DOI: 10.1093/bioinformatics/btl036
The LCB Data Warehouse
Bioinformatics. 22, 8, 1024-6. (2006). DOI: 10.1093/bioinformatics/btl036