Highlighted Projects
Autonomous phenomics for drug/compound profiling
The purpose of this project is to improve studies of mechanisms and pathways for compounds, such as drug or environmental contaminants, with intelligent data generation, automation and AI. To this end we are developing an intelligent system for phenotypic cell profiling (phenomics) that is able to autonomously suggest the next most informative experiment, perform it using an automated lab, learn from the results, and iterate.
The theoretical experimental space of biomedical research is vast. Even when restricting it to e.g. find optimal combinations and concentrations of drugs for treatment, it becomes infeasible to search exhaustively using brute force. As an example, evaluating all combinations of 3 drugs from a panel of 500 would require almost 24 M experiments. This constitutes a...
Phenotypic Drug Screening
The purpose of this project is to apply phenotypic profiling for the discovery of novel or medically available drugs against cancer, with special initial interest on Sarcoma.
Overview
Phenotypic drug discovery (PDD) is one of the two major branches in the drug discovery field, the other one being in vitro targeted screening. One of the main approaches of PDD is phenotypic profiling of cells by high-throughput high-content microscopy. This allows for the identification of morphological changes in the cell that are indicative, for instance, of the mode of action (MoA) of a drug, novel compound or biological. This is typically done by using either brightfield imaging, or applying a coktail of dyes to the cells to stain cellular organelles (e.g. using the Cell Painting protocol)....
AI/Machine Learning with confidence in Drug Discovery
This project aims at developing computational methods, tools and AI models to aid the drug discovery process. We primarily focus on ligand-based methods such as structure-activity relationships (SAR or QSAR). A special emphasis is on achieving a valid measure of confidence in predictions, and we apply Conformal Prediction methodology to this end. In order to analyze large-scale data we make use of modern e-infrastructure such as high-performance computing clusters, cloud computing resources, containerized microservice environments such as Kubernetes, and data analytics platforms such as Apache Spark.
Conformal Prediction
Conformal Prediction (CP) provides a layer on top of existing machine learning methods and produces valid prediction regions for test objects. This contrasts to...
Long-read sequencing for clinical applications
Long-read sequencing is a new technology that offers important advantages for medical diagnostics as compared to the short-read sequencing technologies that are currently dominating the market. We have together with clinicians developed a data analysis product (CLAMP) and implemented the world’s first clinical routine diagnostics of leukemia mutations using long-read sequencing, and are now expanding to other applications.
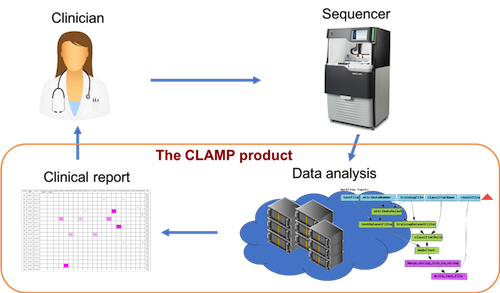

Left: The project aims to provide a complete informatics system for working with long-read sequencing in clinical diagnostics, including an automated analysis module, a searchable database, and a user interface. All components were developed in collaboration with clinicians. Right: The project was selected on the Royal Swedish Academy of Engineering Sciences, IVA...
HASTE: Hierarchical Analysis of Spatial and TEmporal image data
From intelligent data acquisition via smart data-management to confident predictions
The HASTE project takes a hierarchical approach to acquisition, analysis, and interpretation of image data. We develop computationally efficient measurements for data description, confidence-driven machine learning for determination of interestingness, and a theory and framework to apply intelligent spatial and temporal information hierarchies, distributing data to computational resources and storage options based on low-level image features.

HASTE is a collaboration between the Wählby lab (PI), Hellander lab (co-PI), both at the Department of Information Technology, Uppsala University, the Spjuth lab (co-PI) at the Department of Pharmaceutical Biosciences, Uppsala University, the Nilsson lab at the...

