Phenotypic Drug Screening
The purpose of this project is to apply phenotypic profiling for the discovery of novel or medically available drugs against cancer, with special initial interest on Sarcoma.
Overview
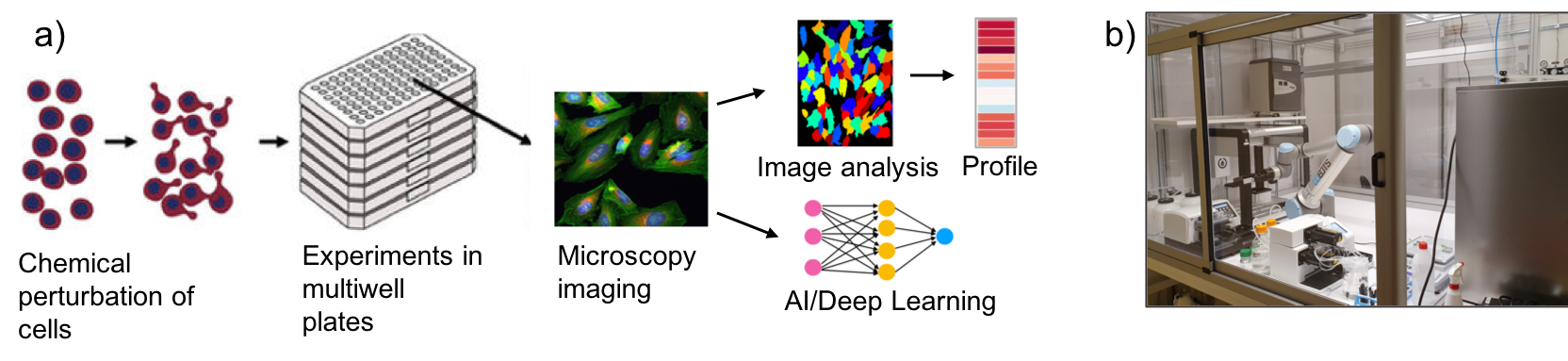
Phenotypic drug discovery (PDD) is one of the two major branches in the drug discovery field, the other one being in vitro targeted screening. One of the main approaches of PDD is phenotypic profiling of cells by high-throughput high-content microscopy. This allows for the identification of morphological changes in the cell that are indicative, for instance, of the mode of action (MoA) of a drug, novel compound or biological. This is typically done by using either brightfield imaging, or applying a coktail of dyes to the cells to stain cellular organelles (e.g. using the Cell Painting protocol).
Morphological changes in the cell can be measured using two major approaches: 1) making use of image analysis software (e.g. Cell Profiler) to identify and segment the cells, and subsequenty extracting selected cellular features, and 2) using Deep Learning (DL) to extract relevant cellular features in an unbiased manner.
Upon the extraction and selection of the morpfhological features, morphological profiles are generated that are unique per each cell type or perturbation. This is achieved through extensive analysis of the vast amount of features extracted at a single cell level.
Aims
In this project we will apply a PDD approach to identify potential novel treatments against Sarcoma:
Aim 1: Morphological profiling of a number of Sarcoma, as well as non-sarcoma/cancer models. We will make use of Cell Painting to morphologically profile different Sarcoma cell lines, and established patient-derived models, as well as non-sarcoma/cancer models, to identify differences among each Sarcoma, classify them, and to correlate this classification with the presence or absence of fusion proteins, which are characteristic of each Sarcoma type.
Aim 2: FDA approved drug screen. We will combine Cell Painting with a library of medically approved drugs, as well as dugs in different Phase trial stages, to identify those that can shift a Sarcoma morphological profile towards a non-sarcoma/cancer profile, and identify potential vulnerabilities.
Aim 3: Validation of the identified FDA approved drugs. We will make use of pre-clinical models to validate the drugs identified in Aim 2.
Methods
We will make use of our atomated lab to carry out the Cell Painting experiments, including the cell work, cell painting protocol, as well as automated imaging, image analysis and data analysis and visualisation.

