NDCP
External Links
NDCP: Non-Disclosed Conformal Prediction
This is an implementation for ligand-based modeling of the method described in:
Aggregating Predictions on Multiple Non-disclosed Datasets using Conformal Prediction
Ola Spjuth, Lars Carlsson, Niharika Gauraha
arXiv:1806.04000 [stat.ML]
Background
It is a common objective for different organizations to be able to contribute to improved predictive models, such as in pre-competitive alliances, but there is often problems with sharing internal data between the parties. One example is in the pharmaceutical industry, where predictions on existing data (such as measured assays of various hazard endpoints for chemical compounds) constitute valuable assets and it would be desirable for all companies to have as good predictive models as possible; but sharing data between companies is usually not so easy.
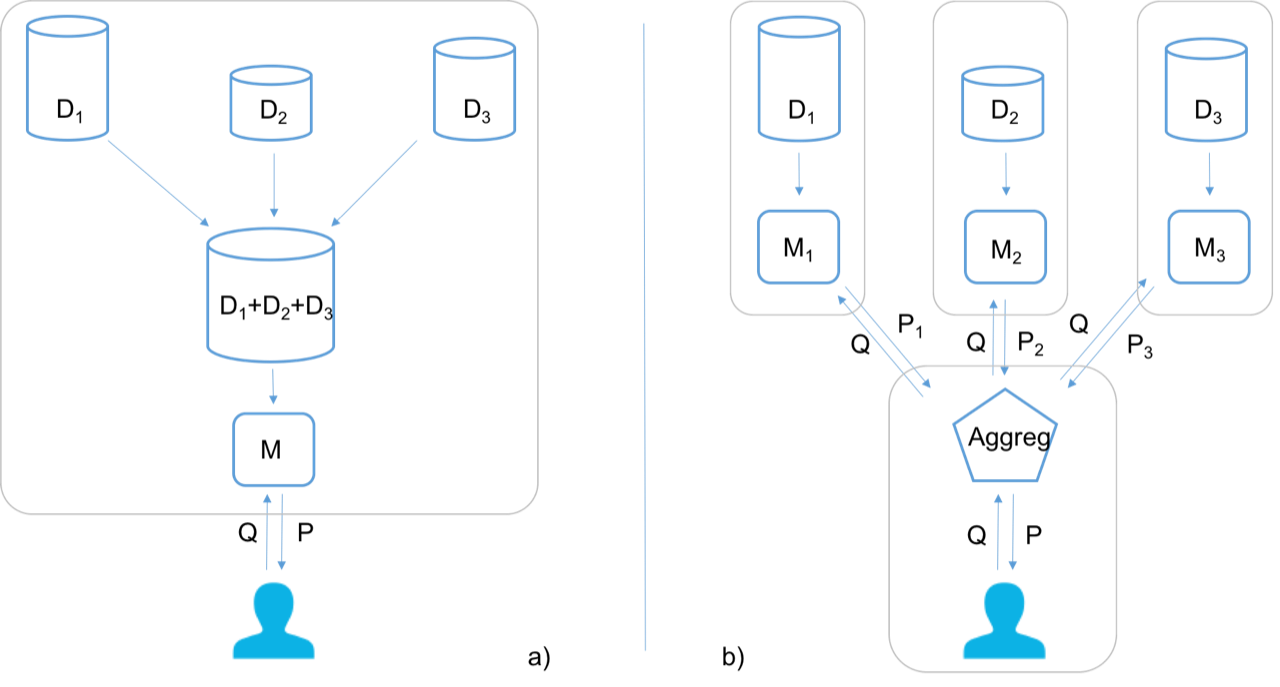
Figure 1: a) The most common approach is to collect data from different data sources (D1 −D3) into a single dataset, which then is used to train a model M that can be used to make a prediction P on a query object Q. b) The aggregated TCP approach implies that a model Mn is trained at each data source Dn, and the query object Q is passed on to each model, and predictions Pn are then aggregated to deliver the resulting prediction P. The gray wireframes are used to visualize the different actors taking part in the procedure independent of each other.
Method
The idea of the method is that the different parties train an individual model of their local data, and make this available as a web service accepting a SMILES as input, and delivers results in form of p-values (one per each class in the classification case), and a prediction interval (in the regression case). No other information is transferred over the network, making the implementation preserve privacy completely. The results from the contributing parties is then merged as described in Spjuth et al 2018 (https://arxiv.org/abs/1806.04000) to yield predictions with lower variance and improved efficiency.
Usage
NDCP is available as two docker container images. One docker image (‘Deploy model’) is deployed on all sites that will serve predictions (without disclosing data); there is no limit to how many different instances that can be deployed. The second docker image (‘Prediction server”) contains a server that aggregates p-values and makes available a web page that can be used to consume the service.
Docker images can be downloaded from:
| Docker image | URL |
|---|---|
| Deploy model | http://pele.farmbio.uu.se/ndcp/deploy-model.tar.gz |
| Prediction server | http://pele.farmbio.uu.se/ndcp/ndcp-predict.tar.gz |
The “model Docker” builds a model from a file with molecules (accepts .sdf or tab separated .tsv with SMILES and property).
Build and deploy model
docker load < deploy-model.tar.gz
Create a data folder mkdir <data dir> and copy SD-file and cpsign-license into the folder.
Some CPSign specific settings are needed:
| Param | Example value | Description |
|---|---|---|
--calibration-ratio |
0.20 |
Part of training set to use for calibration |
--response-name |
activity |
Name of property to model |
--labels |
"A,N" |
Values of property to model |
--cptype |
1 |
Specify that we are doing classification |
--nr-models |
1 |
Number of models to build |
--model-name |
"TestMod" |
Name of the model that is built |
These parameters are added at the end when building, see below:
docker run -d -p 8080:8080 -v <absolute data dir>:/build/source <image name:tag> \
--calibration-ratio 0.20 --nr-models 1 --labels "A,N" --logfile /build/full.log \
--cptype 1 --response-name activity --model-name "TestMod"
Deploy webpage to view aggregation
docker load < ndcp-predict.tar.gz
docker run -p 8080:8083
Running example
A running example is deployed at http://ndcp.service.pharmb.io
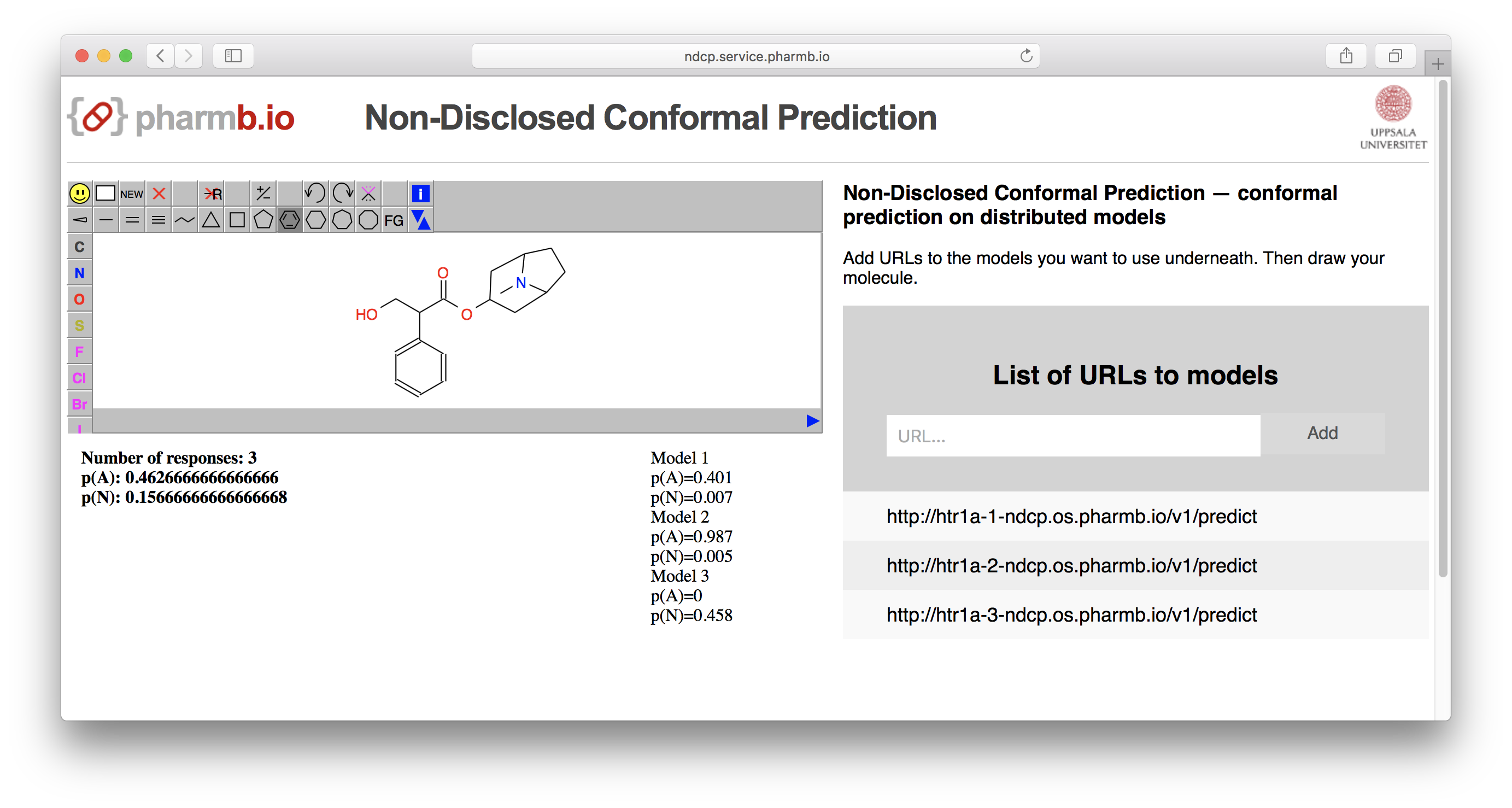
Figure 2: Example of a running deployment of NDCP.
Ligand-based implementation
The implementation uses CPSign (http://cpsign-docs.genettasoft.com/) as the underlying modeling method with SVM and Signatures. A license is required but these are generously provided by GenettaSoft.





