Long-read sequencing for clinical applications
Long-read sequencing is a new technology that offers important advantages for medical diagnostics as compared to the short-read sequencing technologies that are currently dominating the market. We have together with clinicians developed a data analysis product (CLAMP) and implemented the world’s first clinical routine diagnostics of leukemia mutations using long-read sequencing, and are now expanding to other applications.

Left: The project aims to provide a complete informatics system for working with long-read sequencing in clinical diagnostics, including an automated analysis module, a searchable database, and a user interface. All components were developed in collaboration with clinicians. Right: The project was selected on the Royal Swedish Academy of Engineering Sciences, IVA 100 list as a project that has high innovative potential and relevance for resulting in new products and services. Ola Spjuth presented the project at IVA Research2Business conference on March 20, 2019.
Long-read single molecule sequencing (LR-SMS) is often called the “third generation” of DNA sequencing and offers important advantages both compared to the 1st (Sanger-based) and 2nd generation (short-read) sequencing technologies. Our team has been among the first in the world to adapt LR-SMS for clinical applications. One important result from our research is that LR-SMS enables for detection of cancer mutations at an earlier stage as compared to other sequencing technologies, thereby opening up for improved patient treatment and better use of expensive drugs. We have also implemented the world’s first clinical routine diagnostics using LR-SMS, screening for drug resistance mutations in patients diagnosed with Chronic Myeloid Leukemia (CML) and who are not responding to treatment. Over 300 patient samples have been analysed so far, and in several cases we have identified therapeutic resistance mutations below the limit of detection of alternative methods, leading to an improved therapeutic decision support and more personalized treatment.
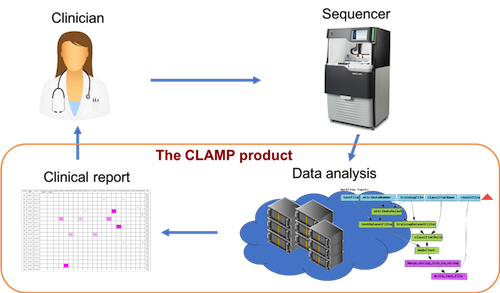
A central part of our project is the software solution for analysis of LR-SMS data. Over the last 6 years we have developed a software named CLAMP through a close collaboration between bioinformaticians, sequencing experts and clinicians. The first application of CLAMP is the screening of BCR-ABL1 resistance mutations in CML, a system that have been developed together with Uppsala University Hospital over a number of years and running in clinical routine since 2015[1-4]. Today, CLAMP is a fully automated system that takes raw LR-SMS data as input and produces conclusive results for actionable cancer mutations. In 2017, the spin-off company Pincer Bio AB was registered to commercialize CLAMP as a clinical decision support software.
Follow-up projects have been initiated for HCV and TP53. We are also part of the joint SeRC-eSSENCE flagship project e-Science for Cancer Prevention and Control (eCPC).
Main collaborators:
Uppsala University:
Ola Spjuth, PhD; and Wesley Schaal, PhD; Department of Pharmaceutical Biosciences and Science for Life Laboratory, Uppsala University
Adam Ameur, PhD; Department of Immunology, Genetics and Pathology (IGP) and National Genomics Institute (NGI) and Science for Life Laboratory
Uppsala University Hospital
Lucia Cavelier, PhD, and Monica Hermansson, MD, PhD; Clinical Genetics division
Karolinska Institutet:
Ola Spjuth, PhD; Department of Medical Epidemiology at Karolinska Institutet and Swedish e-Science Research Centre
Thierry Soussi, MD, Prof.
[1] Ameur et al. Single-Molecule Sequencing: Towards Clinical Applications. Trends in Biotechnology 2019 Jan;37(1):72-85.
[2] Ardui et al. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Research 2018 Mar 16;46(5):2159-2168.
[3] Lodé et al. Single-molecule DNA sequencing of acute myeloid leukemia and myelodysplastic syndromes with multiple TP53 alterations. Haematologica. 2018 Jan;103(1):e13-e16.
[4] Cavelier er al. Clonal distribution of BCR-ABL1 mutations and splice isoforms by single-molecule long-read RNA sequencing. BMC Cancer. 2015 Feb 12;15:45.


