Just published: 'A phenomics approach for antiviral drug discovery' in BMC Biology
3 Jul, 2021
Our latest work “A phenomics approach for antiviral drug discovery” has now been published at BMC Biology!
Jonne Rietdijk, Marianna Tampere, Aleksandra Pettke, Polina Georgiev, Maris Lapins, Ulrika Warpman-Berglund, Ola Spjuth, Marjo-Riitta Puumalainen & Jordi Carreras-Puigvert. A phenomics approach for antiviral drug discovery. BMC Biology 19, 156 (2021). DOI: 10.1186/s12915-021-01086-1
Youtube video describing the paper and the data publishing efforts.
We developed a new method for phenotypic antiviral drug repurposing and discovery, based on morphological profiling of cells. A collaborative and fruitful effort between our group at Uppsala University and Marianna Tampere, Aleksandra Pettke and Marjo Puumalainen from Karolinska Institutet, and SciLifeLab.
When the pandemic hit, as many others, we put our brains to work against COVID-19. What we do in our lab is morphological profiling of cells using CellPainting for characterisation of environmental toxicants, identification of MoA of novel compounds, drug discovery and drug repurposing. So we thought, can we use morphological profiling for antiviral drug discovery?
The answer is yes. We first wanted to know if we could identify a particular morphological profile in virus-infected cells. For this we used Human Coronavirus 229E to infect MRC-5 Human lung fibroblasts. But to detect the virus-infected cells we needed to add an anti-virus antibody to the CellPainting cocktail, so we modified the original CellPainting protocol by replacing MitoTracker with an anti-coronavirus nucleoprotein antibody. And this worked beautifully.
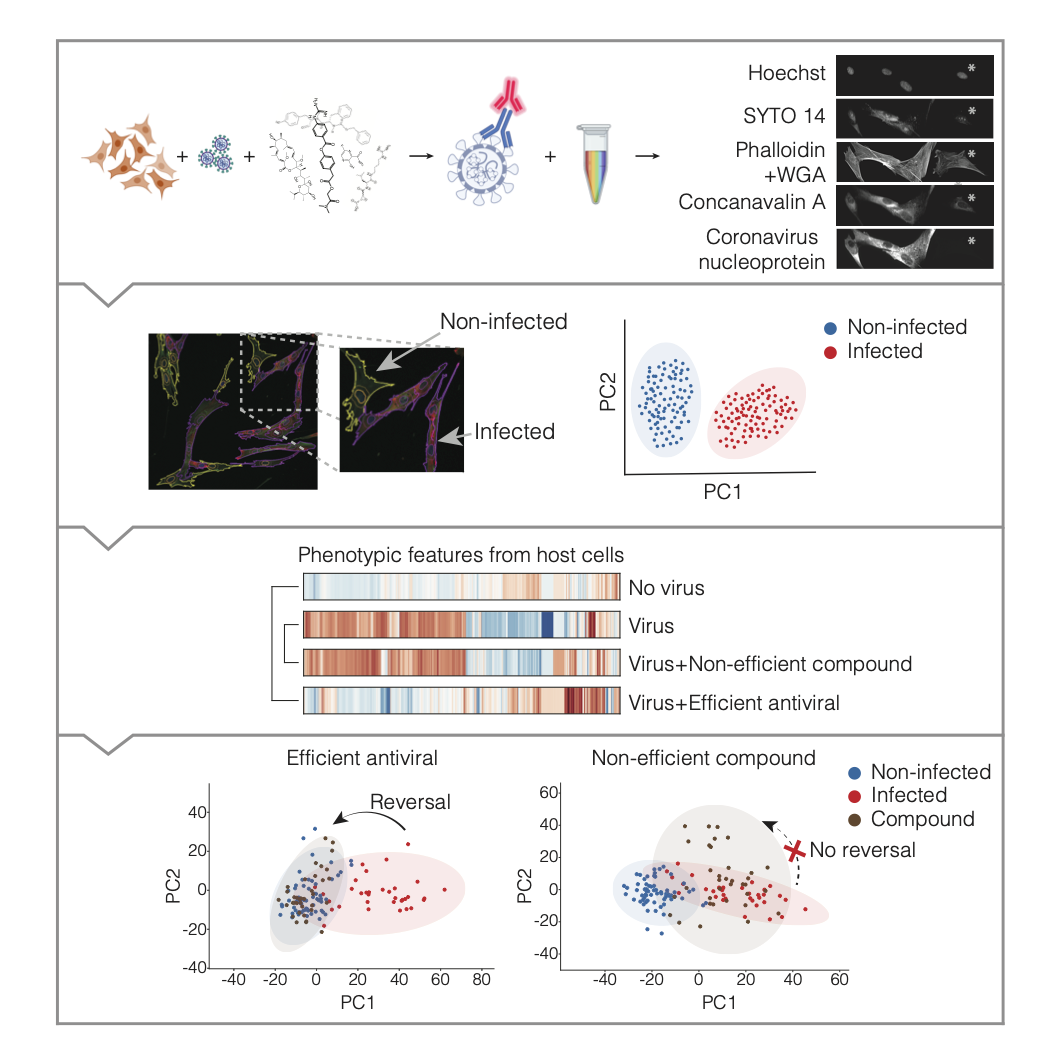
We then designed a Cell Profiler pipeline to identify infected and non-infected cells, and extracted more than 1000 features at a single cell level. At this point, without using the anti-viral antibody, we could clearly see a virus-induced morphological signature. We then had the grounds set for our new method, a phenomics approach to identify antiviral compounds that would reverse the virus-induced phenotype on the infected cells.
We benchmarked our approach, using an antibody-based assay to assess the effect of 9 host and virus-targeting drugs and compounds, out of which 6 are clinically available and 3 were newly synthesised. As previously reported, Remdesivir and E-64d were very efficient antivirals. And we also saw a quite good antiviral effect of the in-house synthesised compounds. Which was very promising and prompted us to later on speculate about how these compounds worked, since their MoA is not fully known yet.
We obtained the morphological signatures of all drugs/compounds in the absence or presence of the virus, and by simply clustering their morphological profiles, and by PCA, we could already see that Remdesivir, E-64d and the 3 novel compounds clustered with the non-infected controls, indicating that these drugs/compounds were reversing the virus-induced phenotype.
One of the advantages of morphological profiling, as it has been shown by others, is that when comparing the profiles of novel drugs to those of a reference set with known targets, one can potentially identify target and MoA of a novel compound. This is of great benefit in drug discovery, specially when performing phenotypic screens. And it let us speculate that the novel compounds might have similar properties than the reference antiviral drugs we used. Of course many more variations are needed.
We think that our new phenomics approach will aid the antiviral drug discovery efforts, and that with only minor adjustments to the image analysis pipelines that we provide, we hope it will be adopted by others to study other disease models and viruses.
The modification that we have done to the CellPainting protocol is just the tip of the iceberg. We imagine many contexts where measuring protein levels with combining an antibody staining with morphological profiling will provide a plethora of data for drawing new hypotheses and pinpointing biological mechanisms.


Jordi and Jonne from the pharmb.io group
We are always advocating for open science, and we have made all the images, data, image analysis pipelines and data analysis code snippets available in the SciLifeLab Data Center repository https://doi.org/10.17044/scilifelab.14188403.v1 and GitHub https://github.com/pharmbio/antiviral-phenomics
This project was made possible through the SciLifeLab National COVID-19 Research Program funded by Knut och Alice Wallenbergs stiftelse.
Press release: https://www.mynewsdesk.com/uu/pressreleases/method-for-discovery-of-antiviral-drugs-3119163
Data Center highlight: https://www.covid19dataportal.se/news/phenomics_method_drug_discovery/

