Autonomous phenomics for drug/compound profiling
The purpose of this project is to improve studies of mechanisms and pathways for compounds, such as drug or environmental contaminants, with intelligent data generation, automation and AI. To this end we are developing an intelligent system for phenotypic cell profiling (phenomics) that is able to autonomously suggest the next most informative experiment, perform it using an automated lab, learn from the results, and iterate.
The theoretical experimental space of biomedical research is vast. Even when restricting it to e.g. find optimal combinations and concentrations of drugs for treatment, it becomes infeasible to search exhaustively using brute force. As an example, evaluating all combinations of 3 drugs from a panel of 500 would require almost 24 M experiments. This constitutes a search space that is infeasible to conquer using conventional methods where experiments and experimental design are carried out by humans. To overcome this challenge, we need intelligent methods for selecting the most informative experiments, integrated with automated lab equipment to perform them. AI-driven, autonomous profiling of cells has the potential to radically speed up biological discoveries, offers a means for detailed understanding of cell states, and allows for testing and optimization of previously impossible treatment strategies.
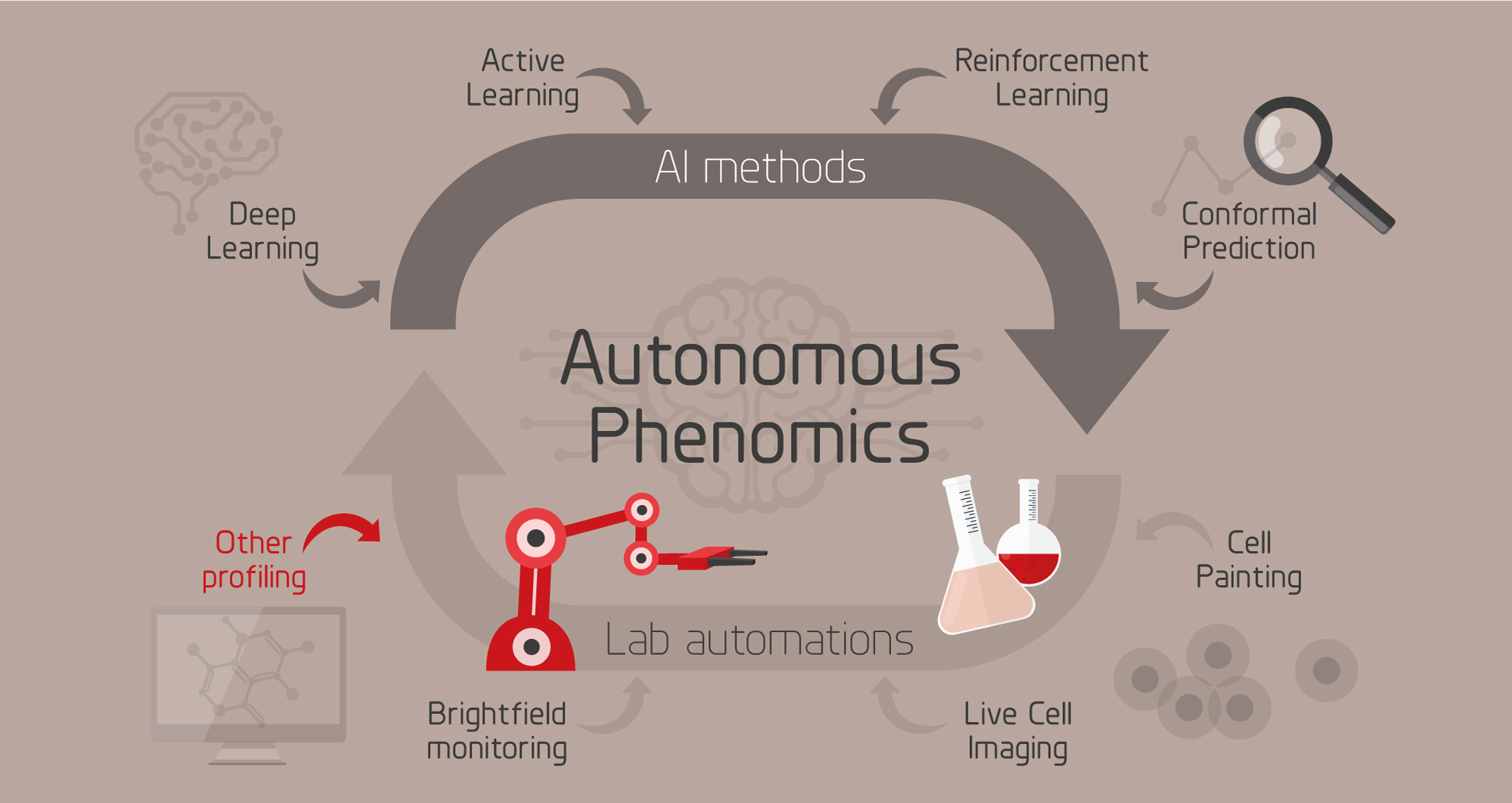
Left: Overview of the autonomous phenomics project. Right: Video from our automated cell profiling lab.
Application 1: Drug treatment optimization in precision cancer medicine
The purpose of this project is to develop an intelligent system for phenotypic cell profiling, being able to autonomously suggest the next most informative experiment, perform it using an automated lab, learn from the results, and repeat. We will apply the developed system to identify novel drug combinations for precision cancer medicine targeting sarcomas.
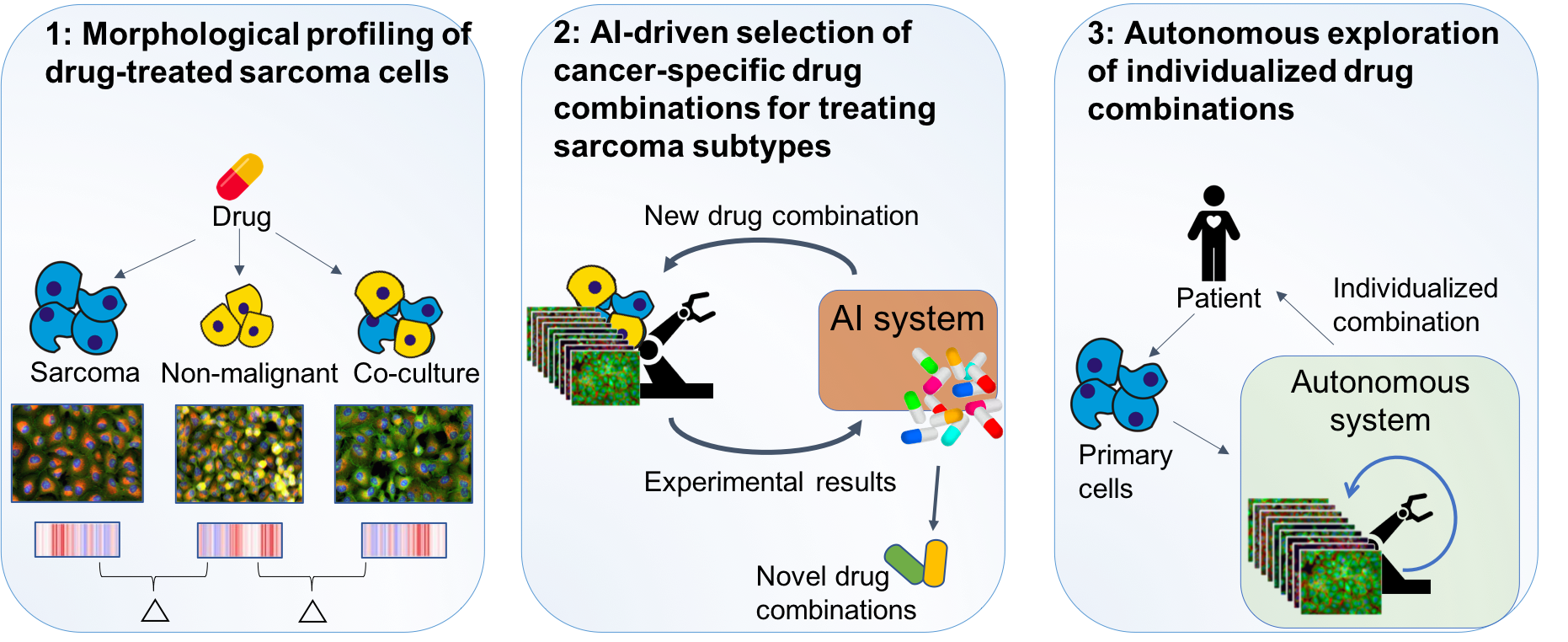
1) We will perform large-scale morphological profiling for individual drug treatments on sarcoma cells alone, non-malignant cells alone, and co-culture of both. The difference in morphologies will contribute to new insights into the biology of sarcomas. The resulting data will also form the basis for 2) where an autonomous system for drug combination treatment will be developed and used to identify novel drug combinations for sarcoma subtypes. In 3) we will apply the developed system to autonomously search for effective individualized combinations towards primary cells from sarcoma patients.
The project is a collaboration with the Kallioniemi lab (precision cancer medicine) and consists of: i) Expansion of our robotized lab into a fully automated cell profiling lab that can be controlled by AI; ii) Development of autonomous AI methods to control the automated lab; iii) Applying the autonomous system to identify optimal drug treatments using cancer cell lines, also in combination with other commonly used drugs to predict positive or negative synergies.
 The project is partially funded by group leader Ola Spjuth receiving a 3.4 MSEK grant from the Swedish Research Council (VR) Natural and engineering sciences, during 2020-2024.
The project is partially funded by group leader Ola Spjuth receiving a 3.4 MSEK grant from the Swedish Research Council (VR) Natural and engineering sciences, during 2020-2024.
Application 2: Autonomous assessment of environmental toxicants
The overall aim of this project is to explore toxicity mechanisms and pathways of environmental contaminants with intelligent data generation, using an automated phenomics laboratory. Current in vitro approaches for this type of research are costly and have long iteration cycles because of manual lab operations and disconnected data analysis, constraining their usefulness. We will develop and optimize a fully automated, robotized cell profiling laboratory with high-content imaging and develop the AI system and supporting informatics infrastructure. The system will design and carry out experiments iteratively and learn with each iteration, and allow us to generate and collect the right data at the right time to answer specific questions related to toxicity mechanisms and pathways of chemicals.
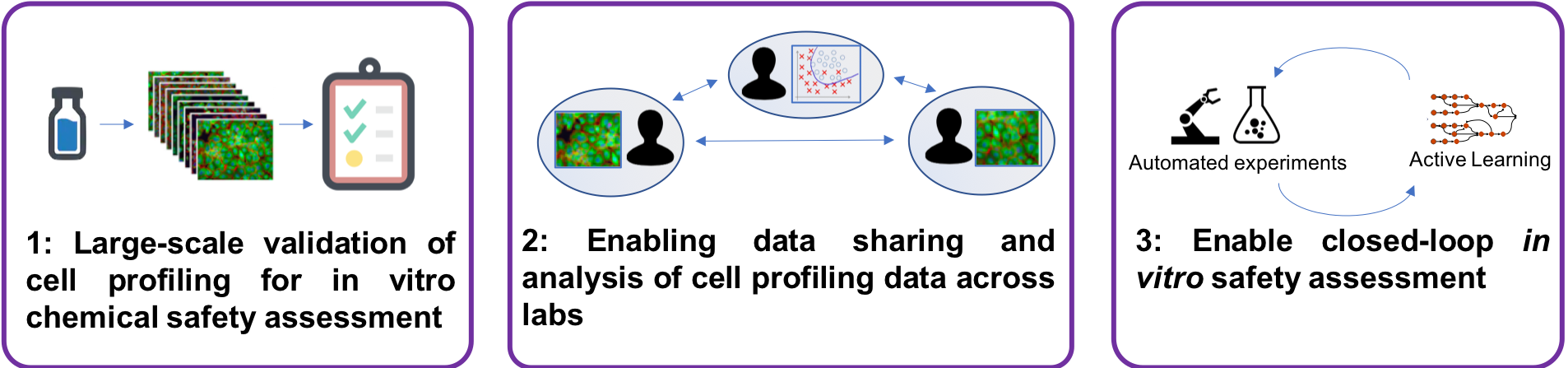
This constitutes a shift from traditional batch- oriented data generation followed by retrospective analysis, to intelligent design of experiments in an online setting where scientific hypotheses are continuously tested and revised in real time as new data is collected. Our project opens up for studying toxicity mechanisms of chemicals on a completely new level and scale, cutting the time and reducing the number of experiments drastically. It will also allow us to explore the synergy between robotized and manual operations for online data generation and collection, a previously unexplored area that has potential to speed up the scientific process.
 The project is partially funded by group leader Ola Spjuth receiving a 3 MSEK grant from the FORMAS Future Research Leaders call during 2019-2022.
The project is partially funded by group leader Ola Spjuth receiving a 3 MSEK grant from the FORMAS Future Research Leaders call during 2019-2022.

